Fig 8.
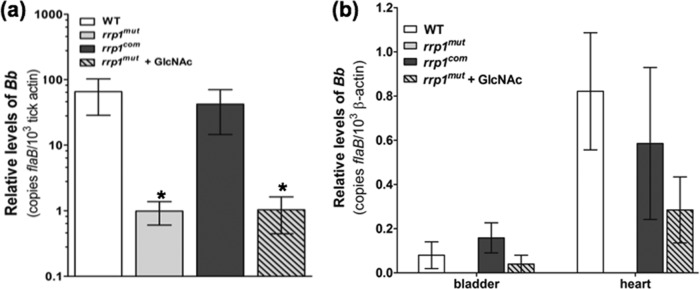
Supplementation of GlcNAc fails to rescue the rrp1mut bacterial load in fed ticks but enables transmission via tick bite. (a) RNA samples were extracted from whole fed ticks (after repletion; 5 to 7 days) and subjected to qRT-PCR analysis. The bacterial burdens in ticks were measured by the number of copies of flaB mRNA compared to the number of copies of tick-actin transcript as previously described (37). The data are presented as the means of relative levels of flaB transcript ± SEM for three groups (each group contained 4 replete ticks) for each strain (WT, rrp1mut mutant, and rrp1com strain). Asterisks indicate that the differences in bacterial load between the WT strain, the rrp1mut mutant, and the rrp1mut mutant supplemented with GlcNAc were statistically significant at a P value of <0.05. (b) Detection of spirochete burdens in C3H mice infected via tick bite using qRT-PCR. At day 14 after tick feeding, mice were sacrificed. Tissues (skin, heart, joint, and bladder) were collected and total RNA from heart and bladder tissues subjected to qRT-PCR analysis as described in Materials and Methods. Statistical analysis showed no significant difference between the WT, the rrp1com strain, and the rrp1mut mutant supplemented with GlcNAc at P = 0.05.
