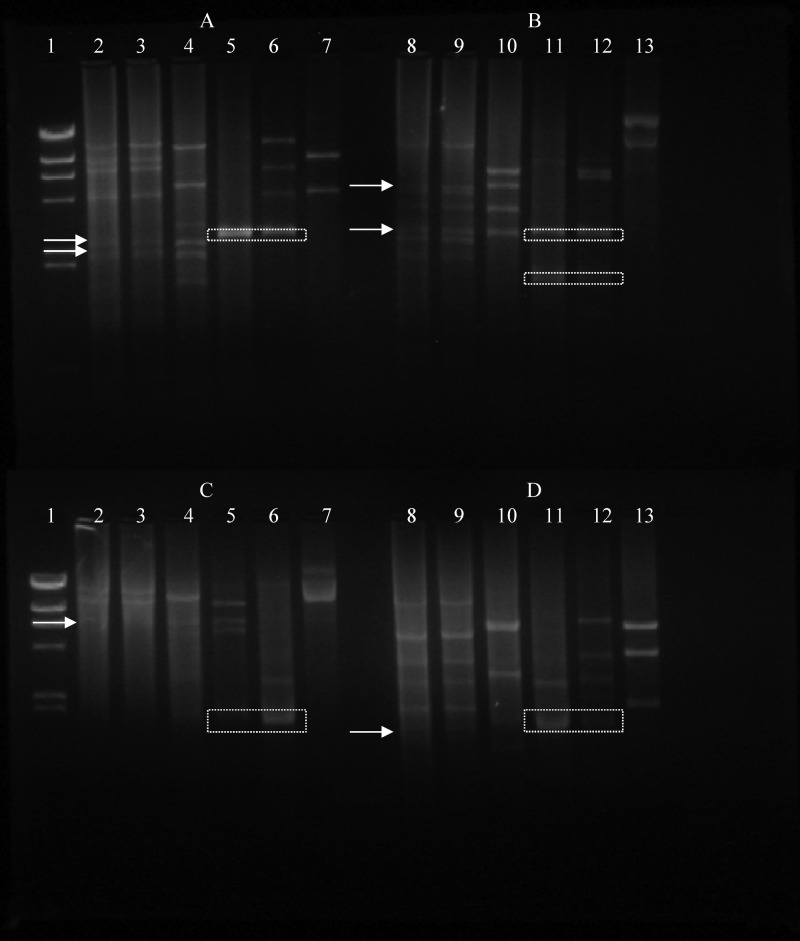
Fig 4.
Plasmid DNA patterns of Lactococcus lactis strains determined using the restriction endonucleases EcoRI (A), EcoRV (B), BamHI (C), and HindIII (D). (A and C) Lanes 1, marker λDNA/HindIII; lanes 2, M78; lanes 3, M104; lanes 4, L. lactis subsp. cremoris LMG 6897T; lanes 5, L. lactis subsp. lactis NCFB 1402 Nis+ strain; lanes 6, L. lactis subsp. lactis F1 Nis+ strain; lanes 7, L. lactis subsp. lactis LMG 6890T. (B and D) Lanes 8, M78; lanes 9, M104; lanes 10, L. lactis subsp. cremoris LMG 6897T; lanes 11, L. lactis subsp. lactis NCFB 1402 Nis+ strain; lanes 12, L. lactis subsp. lactis F1 Nis+ strain; lanes 13, L. lactis subsp. lactis LMG 6890T. Arrows indicate common bands between the L. lactis subsp. cremoris LMG 6897T strain and the wild Nis+ M78 and M104 strains. Squares indicate common bands between the L. lactis subsp. lactis NCFB 1402 Nis+ strain and the L. lactis subsp. lactis F1 Nis+ strain.