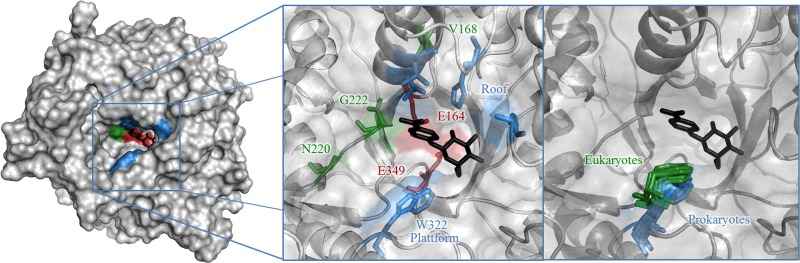
Fig 1.
p-Nitrophenyl-β-d-glucopyranoside docked into the crystal structure of TnBgl1A (atom coordinates are from T. S. Kulkarni, S. Khan, T. Mahmood, A. Sundin, S. Lindahl, C. Turner, D. T. Logan, and E. Nordberg Karlsson, unpublished data). In the left zoomed image, the catalytic residues E164 and E349 are red, the slot platform and roof are blue, and the selected mutation sites are green (except for W322, which is blue). In the right zoomed image, overlaid aglycone platform tryptophan orientations for other family 1 glycoside hydrolases from eukaryotes (Protein Data Bank accession numbers 3AI0, 2E9L, 1E1F, 1V03, and 2RGM) are green and those from prokaryotes (Protein Data Bank accession numbers 1E4I, 1OIN, 1NP2, 3AHX, 1GON, and 1VFF) are blue.