Abstract
The glucose-activated transcription factor carbohydrate response element binding protein (ChREBP) induces the expression of hepatic glycolytic and lipogenic genes. The farnesoid X receptor (FXR) is a nuclear bile acid receptor controlling bile acid, lipid, and glucose homeostasis. FXR negatively regulates hepatic glycolysis and lipogenesis in mouse liver. The aim of this study was to determine whether FXR regulates the transcriptional activity of ChREBP in human hepatocytes and to unravel the underlying molecular mechanisms. Agonist-activated FXR inhibits glucose-induced transcription of several glycolytic genes, including the liver-type pyruvate kinase gene (L-PK), in the immortalized human hepatocyte (IHH) and HepaRG cell lines. This inhibition requires the L4L3 region of the L-PK promoter, known to bind the transcription factors ChREBP and hepatocyte nuclear factor 4α (HNF4α). FXR interacts directly with ChREBP and HNF4α proteins. Analysis of the protein complex bound to the L4L3 region reveals the presence of ChREBP, HNF4α, FXR, and the transcriptional coactivators p300 and CBP at high glucose concentrations. FXR activation does not affect either FXR or HNF4α binding to the L4L3 region but does result in the concomitant release of ChREBP, p300, and CBP and in the recruitment of the transcriptional corepressor SMRT. Thus, FXR transrepresses the expression of genes involved in glycolysis in human hepatocytes.
INTRODUCTION
The liver plays a critical role in maintaining glucose homeostasis, by controlling both glucose production and utilization (1). In the fasting state, the liver produces glucose from glycogen through the glycogenolysis pathway and from lactate, glycerol, and amino acids through the gluconeogenesis pathway (2). In the fed state, energy is provided by glucose oxidation through the glycolysis pathway and glucose excess is stored as glycogen through the glycogen synthesis pathway or converted into fatty acids via the de novo lipogenesis pathway (1). These pathways are regulated not only by glucagon and insulin but, also, by glucose itself.
Glucose regulates its own metabolism by controlling the transcription of glucose-handling enzymes (3). Also known as WBSCR14 or MondoB and member of the basic helix-loop-helix leucine zipper (bHLH-ZIP) family of transcription factors (4), the transcription factor carbohydrate response element binding protein (ChREBP) is necessary for glucose-induced gene expression (5). It associates with the Max-like protein (Mlx) (6) to bind to carbohydrate-response elements (ChOREs) composed of two E-box-like motifs present in the promoters of its target genes, such as the glycolytic liver-type pyruvate kinase (L-PK), lipogenic fatty acid synthase (FAS), and acetyl coenzyme A (acetyl-CoA) carboxylase 1 (ACC1) genes (5, 7, 8). The activation of ChREBP transcriptional activity by glucose involves its dephosphorylation by protein phosphatase 2A (PP2A), allowing its nuclear translocation and binding to ChOREs (9). The generation of xylulose 5-phosphate by the pentose phosphate pathway activates PP2A, hence coupling glucose metabolism to ChREBP activation (10). However, recent reports challenged this view, showing that glucose 6-phosphate mediates ChREBP activation (11, 12). A recent study showed that both xylulose 5-phosphate and glucose 6-phosphate activate ChREBP (13). Moreover, ChREBP activity is also regulated by other posttranslational modifications, such as acetylation (14) and O-GlcNacylation (15, 16). Finally, other transcription factors, including the nuclear receptors hepatocyte nuclear factor 4α (HNF4α) (17) and liver X receptors (LXR) (18), are involved directly or indirectly in the transcriptional response to glucose, offering multiple entry points for the fine tuning of cellular responses to this nutrient.
The nuclear bile acid receptor farnesoid X receptor (FXR) maintains bile acid homeostasis by regulating the expression of key enzymes involved in bile acid synthesis and transport in the liver and intestine (19–22). FXR also controls lipid metabolism, as illustrated by the phenotype of the FXR-deficient (FXR−/−) mouse, which displays elevated serum triglyceride and cholesterol levels (23, 24). More recently, we and others have shown that FXR modulates glucose homeostasis and insulin sensitivity (25–28). Hepatic FXR expression is decreased in rodent models of diabetes and regulated in vitro by insulin and glucose in rodent hepatocytes (29). Moreover, FXR−/− mice display an accelerated hepatic response to high carbohydrate refeeding (30). FXR negatively regulates the expression of several glucose-regulated genes, such as L-PK, FAS, and ACC1, in rodent primary hepatocytes, which led us to hypothesize that FXR could interfere with the transcriptional activity of ChREBP.
In the present study, we show that ligand-activated FXR represses the induction of L-PK gene expression during the fasting-to-high-carbohydrate transition period in livers of wild-type mice. Furthermore, this regulation is conserved in immortalized human hepatocytes (IHH), which display glucose and insulin responsiveness (31). Investigation of the molecular mechanisms by which FXR inhibits the response to glucose shows that FXR inhibits glucose-induced L-PK expression by a transrepressive mechanism involving the release of the transcription factor ChREBP and the recruitment of the transcriptional corepressor SMRT to the ChORE of the L-PK promoter. The expression of several other previously identified ChREBP or glucose target genes is similarly regulated. These results identify a novel regulatory mechanism of glucose-induced genes by the nuclear bile acid receptor FXR, involving the transrepression of ChREBP-controlled pathways.
MATERIALS AND METHODS
In vivo study and blood and tissue sampling.
All studies were approved by the ethical committee. Twelve wild-type mice were purchased from Charles River (Wilmington, MA), housed in a pathogen-free barrier facility with a 12-h light/12-h dark cycle, and maintained on a standard laboratory chow diet (A04 diet; SAFE diets, Augy, France). The fasting-refeeding experiments were performed as described in reference 30. Before feeding the high-carbohydrate diet, the mice were pretreated (30 min) by gavage with vehicle alone (0.5% carboxymethyl cellulose [CMC]–0.1% Tween 80) or 30 mg of INT-747 (chenodeoxycholic acid [6-CDCA]) per kg of body weight. Plasma glucose concentrations were determined by using Glucotrend 2 (Roche Diagnostics). Total RNA was isolated from livers by guanidinium thiocyanate-phenol-chloroform extraction (32).
IHH and HepaRG cell culture, stimulations, and transfections.
IHH and HepaRG cells were cultured and stimulated with 11 mM and 25 mM glucose, respectively, as described previously (31, 33, 34). For some experiments, as indicated below, IHH cells were pretreated for 1 h with trichostatin A (TSA) (300 μg/ml) before incubation with glucose.
IHH cells were transfected with plasmids using Jet PEI (QBiogene, Illkirch, France) in Williams E (Invitrogen, Cergy-Pontoise, France) medium for 16 h and then stimulated with glucose. IHH and HepaRG cells were transfected as previously described (31, 35) with the following small interfering RNAs (siRNAs): OnTargetPlus SmartPools, scramble control (product no. D-001810; Dharmacon, Lafayette, CO), FXR (product no. L-003414; Dharmacon), ChREBP (product no. L-009253; Dharmacon), CBP (product no. L-003477; Dharmacon), and p300 (product no. L-003486; Dharmacon) siRNAs; siGenome SmartPools scramble control (product no. D-001206; Dharmacon); and SMRT siRNA (product no. M-020145; Dharmacon).
Total RNA extraction and quantitative PCR.
Total RNA was extracted from cells with TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Reverse transcription was performed using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Life Technologies, Cergy Pontoise, France) and 1 μg of total RNA. Quantitative PCRs were performed on an Mx3500p apparatus (Agilent Technologies, Santa Clara, CA) by using the SYBR green Brilliant II fast kit (Agilent Technologies). mRNA levels were normalized to the 36B4 gene mRNA levels, and the fold induction was calculated using the cycle threshold (ΔΔCT) method (36).
GST pulldown.
Glutathione-S-transferase (GST) fusion proteins were adsorbed to glutathione-Sepharose 4B beads (GE Healthcare, Chalfont St. Giles, United Kingdom) for 1 h at 4°C in lysis buffer (1× phosphate-buffered saline [PBS], 1 mM dithiothreitol [DTT], 0.1× Complete protease inhibitor cocktail [PIC; Roche Diagnostics, Meylan, France], 0.5 mM benzamidine). Proteins of interest were produced using the T7 or SP6 TNT system (TNT quick coupled transfection/translation kit; Promega, Madison, WI) containing [35S]methionine according to the manufacturer's protocol. GST or GST fusion protein was preincubated with vehicle (dimethyl sulfoxide [DMSO]) or synthetic FXR ligand (GW4064, 5 μM) for 30 min at 20°C in binding buffer (20 mM Tris-HCl, pH 8, 100 mM KCl, 20% glycerol, 0.2 mM EDTA, 0.05% NP-40, 1 mM DTT, 1 mM benzamidine, 2.5 mg/ml bovine serum albumin, 0.1× PIC) and then with 15 μl of in vitro-translated proteins for 2 h at 20°C in the same buffer. The beads were then washed three times with wash buffer (20 mM Tris-HCl, pH 8, 100 mM KCl, 20% glycerol, 0.2 mM EDTA, 0.05% NP-40, 1 mM DTT, 1 mM benzamidine, 0.1× Complete PIC). Bound proteins were separated by 10% SDS-PAGE. The gels were dried, and radiolabeled proteins were detected by phosphorimaging after a 2-day exposure.
Total and nuclear protein extraction, immunoprecipitation, and Western blot analysis.
For analysis of protein expression after gene silencing, total proteins were extracted by lysing cells in a buffer containing 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100 supplemented with 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 μg/ml leupeptin.
For immunoprecipitation experiments, cytoplasmic proteins were extracted in a buffer containing 20 mM HEPES, pH 7.9, 10 mM KCl, 1 mM EDTA, 0.2% NP-40, 0.1× Complete PIC for 5 min at 4°C. Nuclear proteins were then extracted in the same buffer without NP-40 and supplemented with 0.35 M NaCl for 30 min at 4°C. Protein concentrations were determined by the Peterson method, and equal protein amounts were analyzed by Western blot analysis. Nuclear extracts were immunoprecipitated without antibody (Ab) (control) or with the antibodies against FXR (Ab1, PP-A9033A-00 [R&D Systems, Minneapolis, MN]; Ab2, sc-13063 [Santa Cruz]) for 16 h at 4°C and then 2 h at 4°C with protein A-conjugated agarose beads (Millipore, Molsheim, France). Bound proteins were eluted in Laemmli buffer by heating (90°C for 5 min) and then analyzed by Western blotting.
Protein samples were resolved in 10% SDS-PAGE. After protein transfer, nitrocellulose membranes were incubated with primary antibodies against ChREBP (sc-21189; Santa Cruz Biotechnology, Santa Cruz, CA), FXR (PP-A9033A-00; R&D Systems), SMRT (06-891; Millipore), lamin A/C (sc-6215; Santa Cruz), or beta-actin (sc-1616; Santa Cruz) at 4°C for 16 h and then with secondary antibodies for 1 h at room temperature. Proteins were detected using the enhanced-chemiluminescence Western blot detection kit (Amersham ECL plus Western blotting detection system; GE Healthcare).
Chromatin immunoprecipitation (ChIP) experiments.
Cells were fixed using 0.07% ethylene glycol bis-succinimidylsuccinate (EGS) (Pierce, Rockford, IL) for 30 min and 1% formaldehyde (Sigma-Aldrich, St. Quentin Fallavier, France) for 10 min at 20°C. Chromatin fragmentation was performed using the Bioruptor system (Diagenode, Liège, Belgium). Cross-linked DNA-protein complexes (200 μg) were immunoprecipitated with 4 μg of ChREBP (sc-21189 [Santa Cruz] and NB400-135 [Novus Biologicals, Littleton, CO]), HNF4α (sc-8987; Santa Cruz), FXR (equal mix of sc-13063 [Santa Cruz] and ab28676 [Abcam, Paris, France]), CBP (sc-369; Santa Cruz), p300 (sc-584; Santa Cruz), SMRT (06-891; Millipore), and acylated histone H3 lysine 9 (H3K9-Ac) (07-473; Millipore) antibodies or no antibody as a negative control. Complexes were captured with 50 μl of a 50% protein A-agarose bead slurry (Millipore). DNA-protein complexes were eluted with elution buffer (1% SDS, 0.1 M NaHCO3, 200 mM NaCl). DNA-protein cross-linking was reversed by overnight incubation at 65°C. DNA was then purified using a QIAquick PCR purification kit (Qiagen, Courtaboeuf, France). L-PK or TxNIP promoter regions containing the ChORE or myoglobin promoter (negative control) were targeted for amplification by quantitative PCR by using specific primers (L-PK, 5′-AGAGCCTCCCGTGTGTTAAA-3′ [forward] and 5′-GTGTCACCACTGTCTCCTGTTC-3′ [reverse]; TxNIP, 5′-GCCGCTCCAGAGCGCAACAAC-3′ [forward] and 5′-GCCCTCGTGCACATCCCTCCC-3′ [reverse]; and myoglobin, 5′-AGC ATG GTG CCA CTG TGC T-3′ and 5′-GGC TTA ATC TCT GCC TCA TGA TG-3′). Promoter occupancy was calculated on the basis of the difference between amplification obtained after immunoprecipitation and input and using the myoglobin gene promoter region as a negative control.
Microarray analysis.
Total RNA was prepared from IHH cells stimulated by low and high glucose concentrations, as previously described (31), in the presence of DMSO or GW4064 (5 μM) for 24 h. mRNA quality was analyzed on a Bioanalyzer 2100 (Agilent Technologies), and only RNA preparations with high integrity (RNA integrity number [RIN] of >9) were used. RNA was amplified and labeled using the quick amp labeling kit (Agilent Technologies) and hybridized to Agilent whole human genome GE 4x44K microarrays (Agilent Technologies). Analyses were performed using Genespring version 11.0 (Agilent) and DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/).
Statistics.
Statistical significance was analyzed using the unpaired Student's t test. All values are reported as means ± standard deviations. Results with P values of <0.05 were considered significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
RESULTS
FXR activation inhibits glucose-mediated induction of L-PK expression in mouse liver and in human IHH and HepaRG hepatocytes.
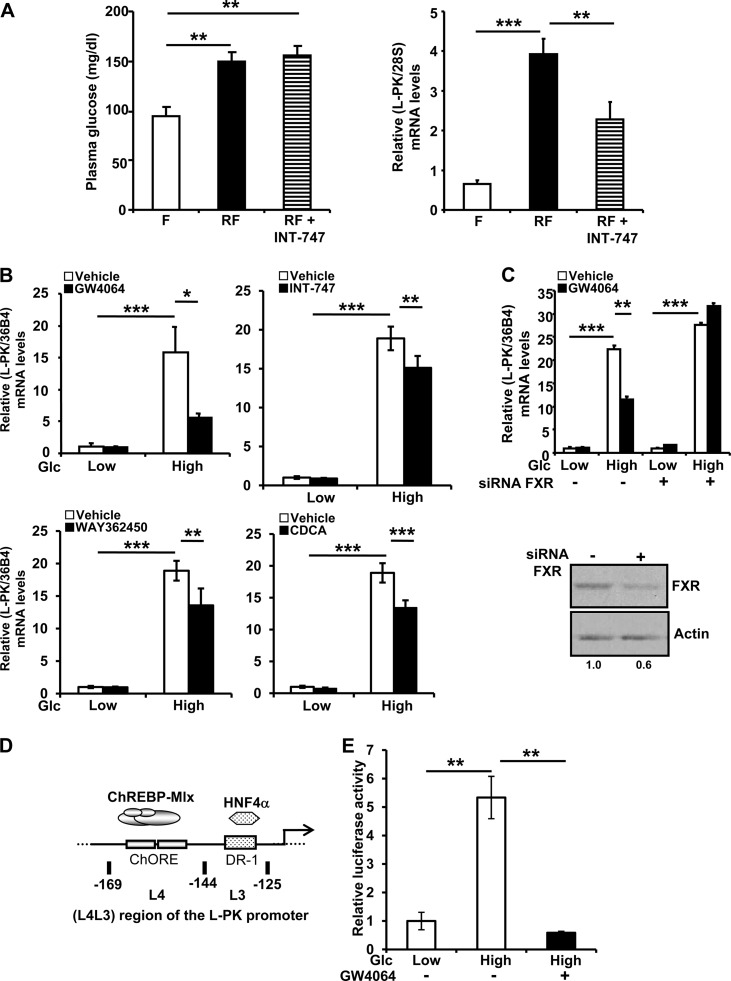
We have previously shown that the glucose-mediated induction of L-PK gene expression depends on FXR in mouse liver (30). To assess the effect of FXR activation on L-PK expression in vivo, wild-type mice were fasted for 24 h, after which they were orally administered INT-747 (6-ECDA, 30 mg/kg), a synthetic FXR agonist (37), and 30 min later, were subjected to a high-carbohydrate diet for 8 h. Plasma glucose levels were significantly induced by high-carbohydrate refeeding (Fig. 1A, left). Hepatic L-PK mRNA levels increased significantly upon high-carbohydrate refeeding, whereas INT-747 treatment significantly repressed this induction (Fig. 1A, right).
Fig 1.
FXR activation inhibits the glucose-induced expression of the L-PK gene in mouse liver and IHH cells. (A) Glycemia (left) and hepatic L-PK mRNA levels (right) in wild-type mice subjected to 24 h of fasting (F) and then refed (RF) for 8 h with a high-carbohydrate diet after being pretreated (30 min) by gavage with vehicle alone (0.5% CMC–0.1% Tween 80) or INT-747 (30 mg/kg). Liver mRNA levels were measured by real-time quantitative PCR. Values are expressed relative to those in the fasting state, arbitrarily set to 1. (B) L-PK mRNA expression in IHH incubated for 24 h in a medium containing low (1 mM) or high (11 mM) glucose concentrations and vehicle (DMSO) or GW4064 (5 μM), WAY362450 (5 μM), INT-747 (10 μM), or chenodeoxycholic acid (CDCA, 100 μM). (C) Effects of FXR silencing on L-PK mRNA (top) and FXR protein (bottom) levels in IHH incubated for 24 h at low (1 mM) or high (11 mM) glucose concentrations and with vehicle (DMSO) or GW4064 (5 μM). L-PK and control 36B4 mRNA levels were measured by real-time quantitative PCR, and values are expressed relative to those at low glucose concentration with vehicle, arbitrarily set to 1. Total protein was extracted and analyzed as indicated in Materials and Methods. FXR protein levels were quantified by densitometry and normalized to the actin protein level. (D) Schematic representation of the L4L3 (−169 to −125) region of the human L-PK promoter that contains the ChREBP (ChORE) and HNF4α (DR-1) binding sites. (E) Activity of the L4L3 region of the L-PK promoter in IHH cells transfected with pGL3-TK-(L4L3)-LPK and incubated for 24 h in medium containing low (1 mM) or high (11 mM) glucose concentrations and vehicle (DMSO) or GW4064 (5 μM). Values are normalized to β-galactosidase activity and are expressed relative to those of cultures with pGL3-TK-(L4L3)-LPK and pCMV-Sport6 (B), pcDNA3 (C), or pGL3-TK-(L4L3)-LPK with vehicle at low glucose concentration (D), which were arbitrarily set to 1.
To test whether FXR activation also affects the induction of glucose-regulated gene expression in human hepatocytes, IHH and HepaRG cells, which respond to glucose and insulin via induction of the glycolytic and lipogenic pathways (31), were incubated at a low glucose concentration (IHH, 1 mM, and HepaRG, 0.5 mM) for 16 h, mimicking the in vivo fasting state, and then switched to a medium containing a high glucose concentration (IHH, 11 mM, and HepaRG, 25 mM) for 24 h in the presence of insulin, mimicking the in vivo high-carbohydrate refed state. IHH cells were treated with the synthetic FXR agonists GW4064 (5 μM), WAY362450 (5 μM), and INT-747 (10 μM), the natural FXR agonist chenodeoxycholic acid (CDCA, 100 μM), or DMSO (vehicle) at the time of high-glucose incubation. HepaRG cells were treated with the synthetic FXR agonist GW4064 (5 μM) or DMSO (vehicle). High glucose concentrations induced L-PK gene expression in IHH (Fig. 1B) and HepaRG (see Fig. S1 in the supplemental material) cells. This induction was strongly blunted upon FXR activation in both cell models. FXR gene knockdown by specific siRNAs in IHH cells, leading to a clear decrease of FXR protein levels (Fig. 1C, bottom), abolished the inhibitory effect of GW4064 (Fig. 1C, top).
IHH cells were then transfected with an expression vector containing the highly conserved L4L3 region of the rat L-PK promoter, containing the ChREBP (ChORE) and HNF4α (DR-1) binding sites involved in glucose induction of L-PK expression (Fig. 1D) (38). L4L3 region-driven promoter activity increased at high glucose concentrations, while FXR activation with GW4064 prevented the glucose-induced activation of the L4L3-driven promoter (Fig. 1E). These results show that the L4L3 region of the L-PK promoter mediates the FXR-dependent inhibition of the glucose-induced response.
FXR does not alter ChREBP nuclear translocation in IHH cells but interacts physically with the ChREBP and HNF4α proteins.
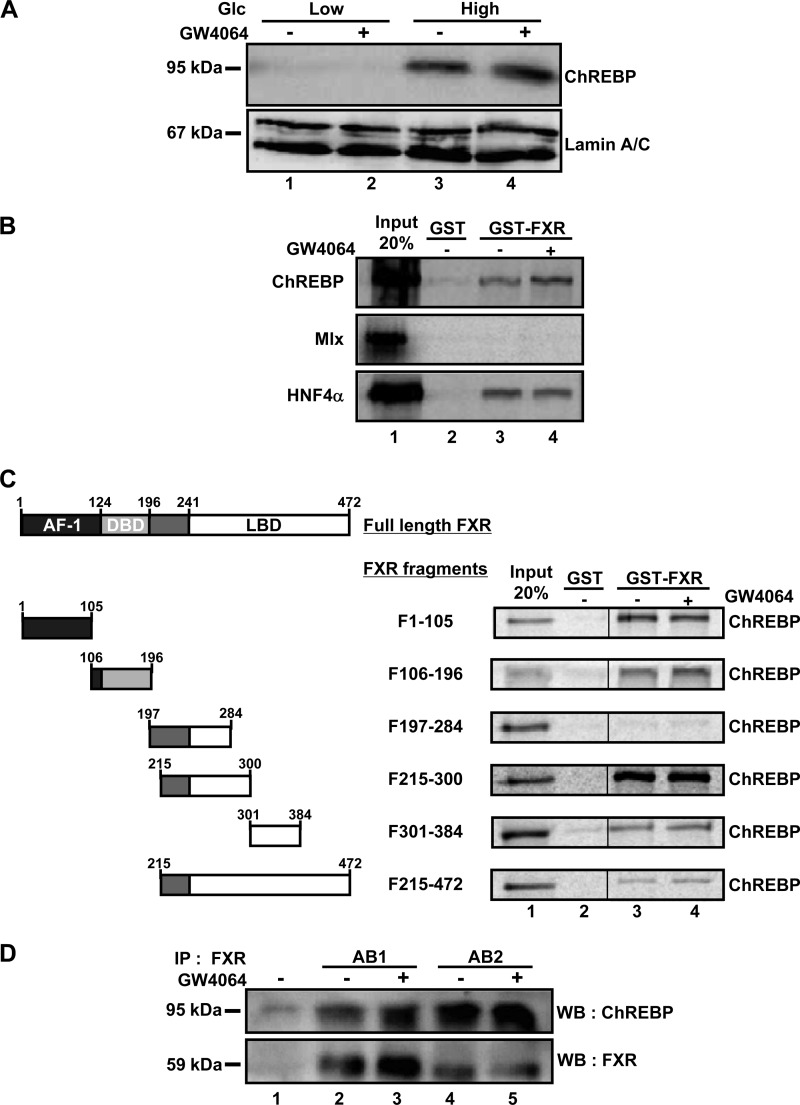
Since the L4L3 region is sufficient to drive FXR inhibition of transcription, we hypothesized that FXR could interfere with the transcriptional activity of ChREBP and/or HNF4α. Indeed, these transcription factors bind to the L4L3 region (39) and are necessary for the induction of L-PK gene expression in IHH cells (35). Moreover, it has been shown that ChREBP and HNF4α interact directly (17). Our first hypothesis was that FXR could interfere with the nuclear translocation of ChREBP. The nuclear expression of the ChREBP protein was examined by Western blot analysis in IHH cells exposed to either low or high glucose concentrations in the presence or absence of GW4064 (Fig. 2A). Nuclear ChREBP protein concentrations were increased at high glucose concentrations compared to their levels at low glucose concentrations. The nuclear enrichment of ChREBP protein was not affected by FXR activation.
Fig 2.
FXR physically interacts with ChREBP and HNF4α in vitro. (A) ChREBP protein levels in nuclear extracts from IHH incubated for 24 h in a medium containing low (1 mM) or high (11 mM) glucose concentrations and vehicle (DMSO) or GW4064 (5 μM). The expression of ChREBP protein was analyzed by Western blotting using a specific antibody. (B) In vitro GST pulldown experiments using full-length GST-FXR and in vitro transcribed/translated (TNT) ChREBP, Mlx, or HNF4α in the presence of [35S]methionine. (C) In vitro GST pulldown experiments using GST (lane 2) and the indicated FXR deletion mutant protein (lanes 3 and 4) and TNT ChREBP in the presence of [35S]methionine and vehicle (DMSO) or FXR ligand (GW4064, 5 μM). AF-1, activation function 1; DBD, DNA binding domain; LBD, ligand binding domain. (D) Two distinct anti-FXR antibodies were used for coimmunoprecipitation of ChREBP in nuclear extracts from IHH cells transfected with pSG5-FXR, pCMV-Sport6-ChREBP, and pcDNA3-Mlx and incubated in a medium containing a high (11 mM) glucose concentration. The expression of FXR and ChREBP proteins was detected 24 h after transfection by Western blotting (WB) using specific antibodies (AB1 and AB2).
We then investigated whether FXR interacts with ChREBP and/or HNF4α. In vitro GST pulldown experiments showed that FXR interacts directly with both ChREBP, in an FXR ligand-stimulated manner (signal intensity, 1.3 with GW4064 versus 1 without GW4064, P < 0.01, n = 12) (Fig. 2B), and HNF4α, in an FXR ligand-independent manner (signal intensity, 1.3 with GW4064 versus 1 without GW4064, P = 0.3, n = 4) (Fig. 2B), but not with Mlx (Fig. 2B). Analysis of FXR deletion mutants revealed that FXR interacts with ChREBP via its activation function 1 (AF-1) domain (FXR fragments F1-105 [nucleotides 1 to 105] and F106-196) and via its ligand binding domain (LBD) (FXR fragment F215-300) (Fig. 2C, lanes 3 and 4). Finally, FXR coimmunoprecipitated with ChREBP in nuclear extracts from IHH cells transfected with ChREBP and FXR and incubated at high glucose concentrations (Fig. 2D). This interaction was observed with two distinct FXR antibodies, Ab1 and Ab2.
FXR activation decreases ChREBP but not HNF4α or FXR binding on the L4L3 region of the L-PK promoter in IHH cells.
To study the complex bound to the L4L3 region of the L-PK promoter, chromatin immunoprecipitation (ChIP) experiments were performed using cross-linked chromatin from IHH incubated at either low or high glucose concentrations and treated with GW4064 or not treated for 5 h. The relative levels of L-PK promoter occupancy by ChREBP, HNF4α, and FXR were measured (Fig. 3). Increasing the glucose concentration enhanced ChREBP and FXR recruitment to the L4L3 region significantly, whereas the occupancy for HNF4α did not change. FXR activation strongly decreased ChREBP binding to the L4L3 region, while HNF4α and, to a lesser extent, FXR proteins were retained on the L4L3 region (Fig. 3).
Fig 3.

FXR activation at high glucose concentrations releases ChREBP but not FXR and HNF4α from the L4L3 region of the L-PK promoter. Relative levels of L-PK promoter occupancy by ChREBP, HNF4α, and FXR on the L4L3 region are shown. The levels of occupancy were evaluated by quantitative PCR in ChIP experiments performed using total extracts from IHH incubated for 5 h in medium containing a low (1 mM) or high (11 mM) glucose concentration and vehicle (DMSO) or GW4064 (5 μM). Occupancies are expressed relative to those at low glucose concentration with vehicle, arbitrarily set to 1. Each experiment was performed at least 3 times, and results are the averages and standard deviations of these experiments.
FXR activation decreases CBP and p300 transcription coactivator binding to the L4L3 region of the L-PK promoter in IHH cells.
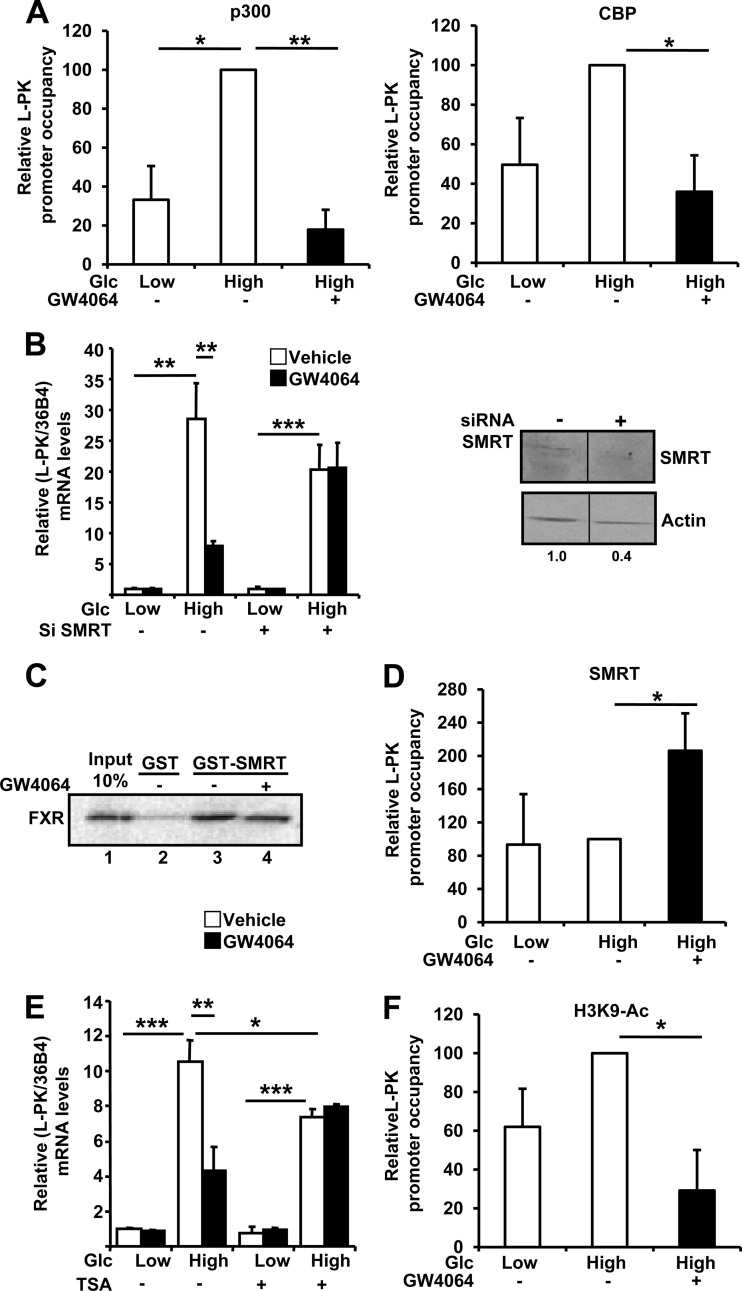
p300 and CBP are transcriptional coactivators associated with ChREBP and/or HNF4α at the regulatory regions of glucose-induced genes in the liver and pancreas (14, 40, 41). Furthermore, p300 and CBP are necessary for the glucose-mediated induction of L-PK gene expression, as shown by the inhibition of the glucose response after p300 and/or CBP silencing (see Fig. 2 in the supplemental material). ChIP experiments showed a significant enrichment of p300 and a trend to increased occupation of CBP on the L-PK promoter at high glucose concentrations in IHH cells (Fig. 4A). Strikingly, FXR activation resulted in the release of both coactivators from the L-PK promoter (Fig. 4A).
Fig 4.
FXR activation leads to the release of coactivators p300 and CBP and the recruitment of the coinhibitor SMRT on the L4L3 region at high glucose concentration. (A) Relative levels of L-PK promoter occupancy by p300 and CBP on the L4L3 region. (B) Effects of SMRT gene silencing on L-PK mRNA (left) and SMRT protein (right) levels in IHH transfected with specific siRNAs and incubated for 24 h at low (1 mM) or high (11 mM) glucose concentration and with vehicle (DMSO) or GW4064 (5 μM). Proteins were extracted and analyzed as indicated in Materials and Methods. SMRT protein levels were quantified by densitometry and normalized to actin protein level. (C) In vitro GST pulldown experiments using full-length GST-SMRT and TNT FXR in the presence of [35S]methionine. (D) Relative levels of L-PK promoter occupancy by SMRT. (E) Effects of TSA treatment on L-PK gene expression in IHH incubated for 24 h at low (1 mM) or high (11 mM) glucose concentration and with vehicle (DMSO) or GW4064 (5 μM). (F) Relative levels of H3K9 acetylation of L-PK. For the experiments whose results are shown in panels A, D, and F, the occupancies were evaluated by quantitative PCR in ChIP experiments performed using total extracts from IHH incubated for 5 h in a medium containing low (1 mM) or high (11 mM) glucose concentrations and vehicle (DMSO) or GW4064 (5 μM). Occupancies are expressed relative to those at low glucose concentration with vehicle, arbitrarily set to 1. Each experiment was performed at least 3 times, and the results are the averages and standard deviations of these experiments. For the experiments whose results are shown in panels B (left) and E, L-PK and control 36B4 mRNA levels were measured by real-time quantitative PCR. The values are expressed relative to those at low glucose concentration with vehicle, which were arbitrarily set to 1.
FXR activation induces the recruitment of the transcriptional corepressor SMRT and leads to the deacetylation of histone H3 lysine 9.
siRNA-mediated silencing of the expression of the transcriptional corepressor SMRT (Fig. 4B, right) in IHH cells did not interfere with glucose-induced L-PK expression but did totally prevent the FXR-dependent repression of L-PK expression at high glucose concentrations (Fig. 4B, left). In vitro GST pulldown experiments showed that FXR interacts directly with SMRT protein (Fig. 4C, lanes 3 and 4). Furthermore, SMRT occupancy of the L4L3 region of the L-PK promoter was not modulated by increasing glucose concentrations but was significantly enhanced upon FXR activation (Fig. 4D). Of note, corepressor NCoR was not recruited on the L4L3 region of the L-PK promoter and silencing of its gene expression did not interfere with the FXR-dependent repression of L-PK gene expression (data not shown).
SMRT recruits and activates histone deacetylase 3 (HDAC3) (42), which removes acetyl groups from histones. IHH cells were thus pretreated for 1 h with TSA, a selective HDAC inhibitor, before being incubated for 24 h in medium containing low and high glucose concentrations supplemented or not with GW4064. Whereas the glucose-induced L-PK expression was diminished by TSA treatment, the FXR ligand-dependent inhibition was totally prevented (Fig. 4E). Moreover, ChIP experiments showed a trend for increased acetylation of histone H3 lysine 9 (H3K9-Ac) on the L-PK promoter in high glucose concentrations, which was significantly inhibited upon FXR activation (Fig. 4F). Taken together, these data suggest that FXR recruits SMRT to the L-PK promoter, leading to histone deacetylation and repression of promoter activity.
FXR inhibits glucose induction of several glucose-induced genes.
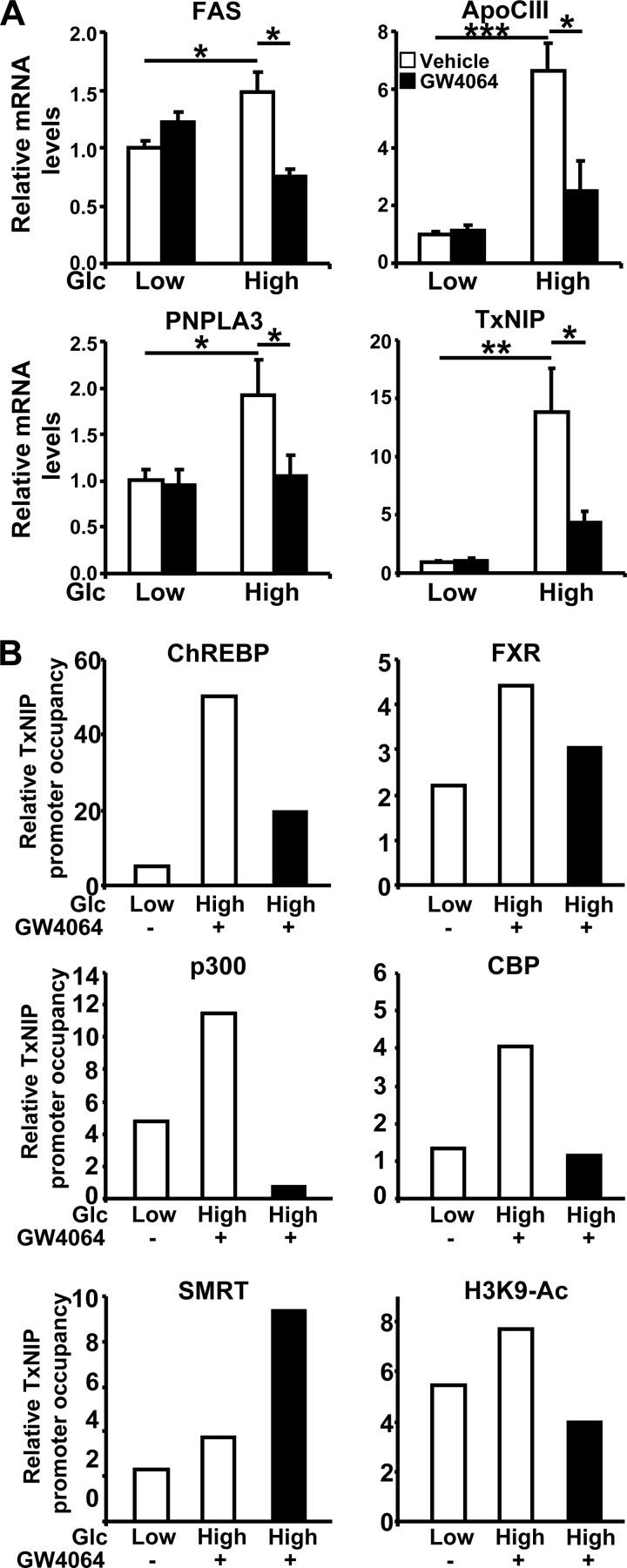
To investigate whether FXR activation generally inhibits the expression of other glucose-induced genes, the levels of expression of known ChREBP target genes, such as FAS (7) and the genes for apolipoprotein (Apo) CIII (35), patatin-like phospholipase domain-containing 3 (PNPLA3) (43), and thioredoxin-interacting protein (TxNIP) (44), were measured. The expression of these genes, which were induced by high glucose concentrations, albeit to differing extents, was consistently inhibited upon FXR activation (Fig. 5A). To investigate whether the same type of molecular mechanism could be involved in the FXR-mediated regulation of these genes, the occupancy of the region around the ChORE binding site of the TxNIP promoter was analyzed using ChIP experiments (Fig. 5B). ChREBP, FXR, p300, and CBP were recruited to the TxNIP promoter at high glucose concentrations. FXR activation resulted in the release of ChREBP, p300, and CBP, with little change in FXR. Moreover, FXR activation led to the recruitment of SMRT, while the acetylation of H3K9 decreased. These results suggest that the activation of FXR inhibits the expression of several ChREBP target genes, probably by a mechanism similar to that observed for the L-PK gene.
Fig 5.
The expression of other ChREBP-regulated genes is also inhibited by FXR by involving a molecular mechanism similar to that for L-PK. (A) mRNA expression of ChREBP target genes in IHH incubated for 24 h in a medium containing low (1 mM) or high (11 mM) glucose concentrations and vehicle (DMSO) or GW4064 (5 μM). Gene and control 36B4 mRNA levels were measured by real-time quantitative PCR. Values are expressed relative to those measured at low glucose concentration with vehicle, arbitrarily set to 1. (B) Relative levels of TxNIP promoter occupancy by ChREBP and FXR (top), p300 and CBP (middle), and SMRT and H3K9 (bottom). The occupancies were evaluated by quantitative PCR amplification of the region of the TxNIP promoter that contains the ChORE in ChIP experiments performed using total extracts from IHH incubated for 5 h at low (1 mM) or high (11 mM) glucose concentrations and vehicle (DMSO) or GW4064 (5 μM). Occupancies are expressed relative to those at low glucose concentration with vehicle, arbitrarily set to 1. Each experiment was performed at least three times, and results from a representative experiment are shown.
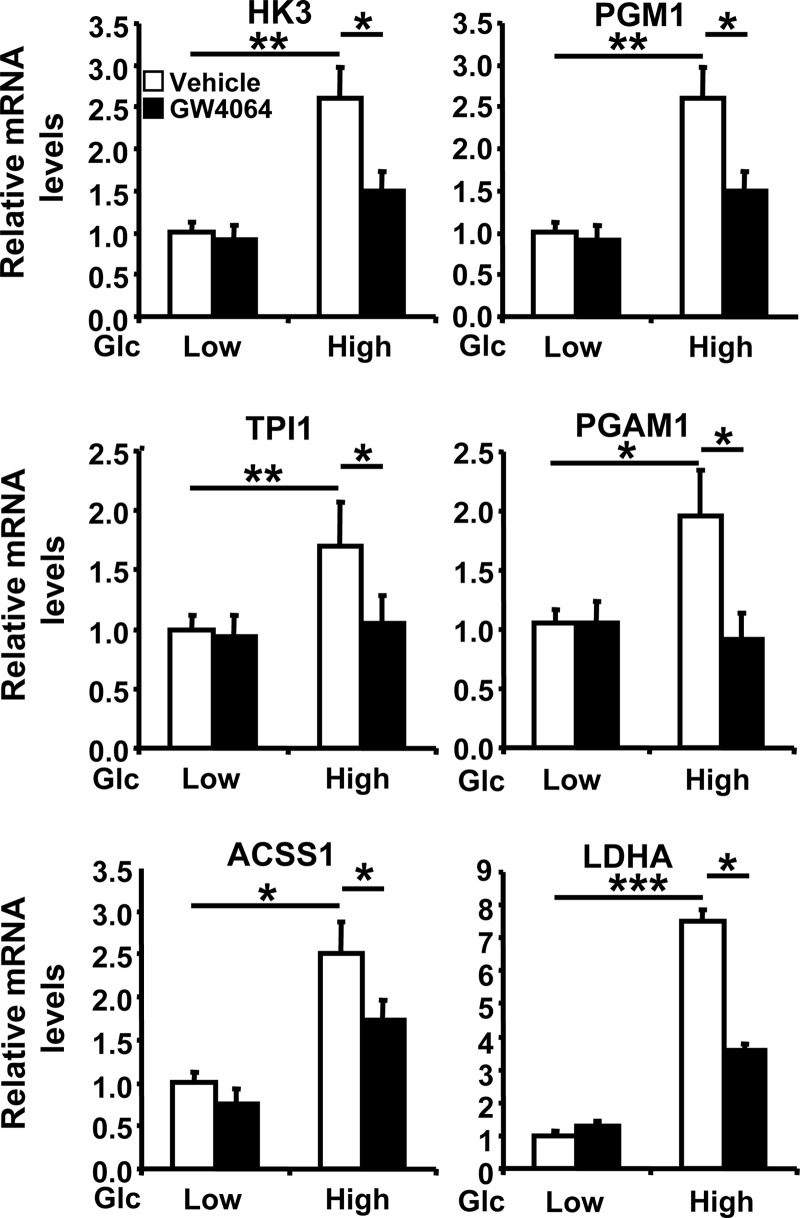
To map the other glucose-induced genes whose expression is inhibited by FXR, a microarray experiment was performed on mRNA isolated from IHH cells cultured in low glucose concentrations for 16 h, followed by another 24 h in either low or high glucose concentrations in the presence or absence of GW4064. Gene array analysis with DAVID Bioinformatic Database software identified a gene cluster that contained six genes coding for enzymes of metabolic pathways involved in glucose utilization and whose expression is regulated both by glucose and FXR (see Fig. S3 in the supplemental material). The expression of these genes was induced at high glucose concentrations in IHH cells (Fig. 6) in a ChREBP- and HNF4α-dependent (see Fig. S4 and S5, respectively, in the supplemental material) manner. Furthermore, the glucose-mediated induction was inhibited upon FXR activation (Fig. 6). Thus, FXR-mediated inhibition seems to be a general phenomenon affecting several ChREBP-regulated genes.
Fig 6.
FXR inhibits the expression of glucose-induced genes. Genes identified as being regulated by both glucose and FXR using microarray analysis were analyzed for mRNA expression in IHH incubated for 24 h in a medium containing low (1 mM) or high (11 mM) glucose concentrations and vehicle (DMSO) or GW4064 (5 μM). HK3, hexokinase 3; PGM1, phosphoglucomutase 1; TPI1, triosephosphate isomerase 1; PGAM1, phosphoglycerate mutase 1; ACSS1, acyl-CoA synthetase short-chain family member 1; LDHA, lactate dehydrogenase A. Gene and control 36B4 mRNA levels were measured by real-time quantitative PCR. The values are expressed relative to those at low glucose concentration with vehicle, which were arbitrarily set to 1.
DISCUSSION
We report here a novel molecular mechanism of transrepression of glucose-induced gene expression by the nuclear receptor FXR. This mechanism was illustrated using two glucose-responsive human hepatocyte cell lines, IHH and HepaRG. This FXR-dependent regulation is mediated by the highly conserved L4L3 region of the L-PK promoter, a region that binds the transcription factors ChREBP and HNF4α and is necessary for glucose induction of L-PK gene expression. FXR does not interfere with the nuclear translocation of ChREBP but does interact with the ChREBP and HNF4α proteins in vitro. High glucose concentrations enhanced the recruitment of ChREBP, p300, CBP, and, surprisingly, FXR but not HNF4α to the L4L3 region, whereas FXR activation led to the release of ChREBP. In parallel, the transcription coactivators p300 and CBP were released, while the transcription corepressor SMRT was recruited. Similar results, with the exception of HNF4α, which does not regulate this gene, were observed for TxNIP, another ChREBP target gene (44). Finally, microarray experiments performed using IHH cells allowed the identification of six genes involved in glucose utilization pathways whose expression is regulated in a similar manner by ChREBP and FXR.
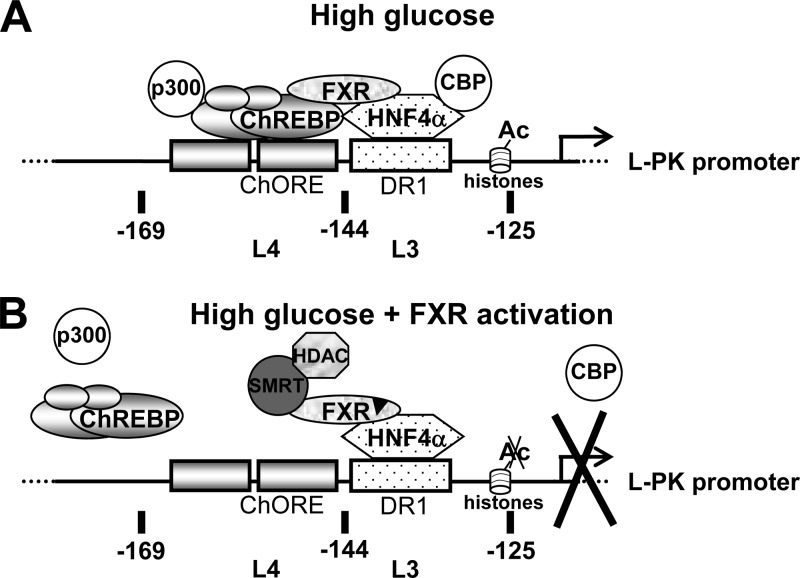
The glucose response is mainly mediated by the transcription factor ChREBP (5), in association with Mlx (6) and in cooperation with other transcription factors, among which can be HNF4α in the case of L-PK (39), FAS (17), and apoCIII (35) but not TxNIP. The transcription factor HNF4α is a master regulator of liver-specific genes (45), regulating the expression of genes involved in glucose utilization (glycolysis) and production (gluconeogenesis) (46). Our results allow us to propose a model of transrepression by the nuclear receptor FXR, involving ChREBP and, in the case of L-PK, HNF4α (Fig. 7). At high glucose concentrations, ChREBP and HNF4α are bound to their binding sites in the L4L3 region of the L-PK promoter and activate L-PK expression through the recruitment of the transcriptional coactivators p300 and CBP (Fig. 7A). Surprisingly, the presence of FXR in this protein complex is induced by high glucose concentrations, likely via a ligand-independent mechanism, possibly involving posttranslational modifications (47). Upon FXR ligand activation, ChREBP is released from the promoter at high glucose concentrations, although HNF4α is still bound, as well as FXR, probably through its interaction with HNF4α (Fig. 7B). Moreover, p300 and CBP are released, whereas the transcriptional corepressor SMRT and HDACs are recruited, thus establishing a repressed transcriptional state. The active recruitment of SMRT by a liganded nuclear receptor contrasts with its known preferential interaction with unliganded nuclear receptors (42) but is in line with the proposed mechanism of transrepression by peroxisome proliferator-activated receptor gamma (PPARγ) (48). Interestingly, FXR appears to participate in both the glucose induction and the inhibition of L-PK gene expression, which suggests a dual role for this nuclear receptor in L-PK regulation. Similar regulation by transrepression was described for other nuclear receptors, such as the glucocorticoid receptor (GR) and PPARα and -γ, which transrepress inflammation pathways (49). To our knowledge, this is the first transrepression mechanism acting on metabolic pathways.
Fig 7.
Model of transrepression of glucose-induced L-PK gene expression by FXR. (A) At high glucose concentrations without FXR activation, ChREBP and HNF4α are bound on the L4L3 region of the L-PK promoter and trans activate gene expression, in part due to the recruitment of the transcriptional coactivators p300 and CBP. FXR is integrated into this protein complex, probably through its direct interaction with ChREBP and HNF4α. (B) At high glucose concentration after FXR activation, ChREBP, as well as p300 and CBP, is released from the L-PK promoter. FXR and HNF4α are still bound on the L-PK promoter. Tethered to the promoter through its interaction with HNF4α, FXR recruits transcriptional coinhibitor SMRT and represses the transcription through the recruitment of HDACs and deacetylation of H3 histones.
Interestingly, the expression of other ChREBP target genes also is regulated negatively by FXR and, likely, by involving a molecular mechanism similar to that for the L-PK gene, at least for FAS, apoCIII, and PNPLA3, whose regulation by glucose is both ChREBP and HNF4α dependent (see Fig. S5 in the supplemental material) (17, 35, 43). On the other hand, the glucose-mediated regulation of TxNIP is ChREBP dependent (44) but not HNF4α dependent (data not shown). Moreover, using cDNA microarray analysis, we have identified six genes involved in metabolic pathways of glucose utilization, glycolysis and expression of which were both glucose induced and FXR inhibited in IHH cells. Except for the lactate dehydrogenase gene (50), these genes have not been previously shown to be ChREBP target genes. We show that the glucose-mediated induction of their expression is ChREBP dependent for all (see Fig. S4 in the supplemental material) and HNF4α dependent for at least five of them (see Fig. S5). Finally, we used data from ChIP sequencing (ChIP-seq) experiments performed on mouse liver (available on the UCSC Genome Bioinformatics website) to localize the peaks of FXR enrichment on the promoters of these genes, and the DNA sequences 30 kb before and 30 kb after every FXR enrichment peak were analyzed to identify potential ChOREs and FXR response elements (FXREs) (see Table S1 in the supplemental material). The DNA regions around approximately 50% of the FXR enrichment peaks do not contain classical FXRE sequences, which suggests that FXR could be recruited to the promoter without direct binding on DNA. Among these regions, one-third contained a ChORE, suggesting a molecular mechanism of regulation like that for the L-PK and TxNIP genes. This percentage is likely an underestimation, since the ChIP-seq data available were obtained on livers of fasting mice. The other regions contained no ChORE, which suggests that FXR could also transregulate gene expression through interaction with an unidentified transcription factor bound to the DNA. These analyses open new perspectives about the identity of FXR-regulated genes, the molecular mechanisms involved, and the functions of FXR.
In summary, our results lead us to propose an original model of the regulation of FXR target gene expression by transrepression and interference with ChREBP transcriptional activity. This new molecular mechanism enhances the understanding of the regulation of glucose handling and lipogenesis by ChREBP and its modulation by FXR.
Supplementary Material
ACKNOWLEDGMENTS
C. Gheeraert and E. Vallez are thanked for their technical help.
This work was supported by grants from the EU Grant HEPADIP (no. 018734), the Region Nord-Pas-de-Calais/FEDER, the Agence Nationale de la Recherche (no. 11 BSV1 032 01), and European Genomic Institute for Diabetes (no. ANR-10-LABX-46). B. Staels is a member of the Institut Universitaire de France.
Footnotes
Published ahead of print 25 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01004-12.
REFERENCES
- 1. Bouché C, Serdy S, Kahn CR, Goldfine AB. 2004. The cellular fate of glucose and its relevance in type 2 diabetes. Endocr. Rev. 25:807–830 [DOI] [PubMed] [Google Scholar]
- 2. Nordlie RC, Foster JD, Lange AJ. 1999. Regulation of glucose production by the liver. Annu. Rev. Nutr. 19:379–406 [DOI] [PubMed] [Google Scholar]
- 3. Girard J, Ferré P, Foufelle F. 1997. Mechanisms by which carbohydrates regulate expression of genes for glycolytic and lipogenic enzymes. Annu. Rev. Nutr. 17:325–352 [DOI] [PubMed] [Google Scholar]
- 4. de Luis O, Valero MC, Jurado LA. 2000. WBSCR14, a putative transcription factor gene deleted in Williams-Beuren syndrome: complete characterisation of the human gene and the mouse ortholog. Eur. J. Hum. Genet. 8:215–222 [DOI] [PubMed] [Google Scholar]
- 5. Yamashita H, Takenoshita M, Sakurai M, Bruick RK, Henzel WJ, Shillinglaw W, Arnot D, Uyeda K. 2001. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc. Natl. Acad. Sci. U. S. A. 98:9116–9121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ma L, Tsatsos NG, Towle HC. 2005. Direct role of ChREBP. Mlx in regulating hepatic glucose-responsive genes. J. Biol. Chem. 280:12019–12027 [DOI] [PubMed] [Google Scholar]
- 7. Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. 2004. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc. Natl. Acad. Sci. U. S. A. 101:7281–7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ishii S, Iizuka K, Miller BC, Uyeda K. 2004. Carbohydrate response element binding protein directly promotes lipogenic enzyme gene transcription. Proc. Natl. Acad. Sci. U. S. A. 101:15597–15602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kawaguchi T, Takenoshita M, Kabashima T, Uyeda K. 2001. Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proc. Natl. Acad. Sci. U. S. A. 98:13710–13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kabashima T, Kawaguchi T, Wadzinski BE, Uyeda K. 2003. Xylulose 5-phosphate mediates glucose-induced lipogenesis by xylulose 5-phosphate-activated protein phosphatase in rat liver. Proc. Natl. Acad. Sci. U. S. A. 100:5107–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dentin R, Tomas-Cobos L, Foufelle F, Leopold J, Girard J, Postic C, Ferré P. 2012. Glucose 6-phosphate, rather than xylulose 5-phosphate, is required for the activation of ChREBP in response to glucose in the liver. J. Hepatol. 56:199–209 [DOI] [PubMed] [Google Scholar]
- 12. Li MV, Chen W, Harmancey RN, Nuotio-Antar AM, Imamura M, Saha P, Taegtmeyer H, Chan L. 2010. Glucose-6-phosphate mediates activation of the carbohydrate responsive binding protein (ChREBP). Biochem. Biophys. Res. Commun. 395:395–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Diaz-Moralli S, Ramos-Montoya A, Marin S, Fernandez-Alvarez A, Casado M, Cascante M. 2012. Target metabolomics revealed complementary roles of hexose- and pentose-phosphates in the regulation of carbohydrate-dependent gene expression. Am. J. Physiol. Endocrinol. Metab. 303:E234–E242 [DOI] [PubMed] [Google Scholar]
- 14. Bricambert J, Miranda J, Benhamed F, Girard J, Postic C, Dentin R. 2010. Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J. Clin. Invest. 120:4316–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guinez C, Filhoulaud G, Rayah-Benhamed F, Marmier S, Dubuquoy C, Dentin R, Moldes M, Burnol A, Yang X, Lefebvre T, Girard J, Postic C. 2011. O-GlcNAcylation increases ChREBP protein content and transcriptional activity in the liver. Diabetes 60:1399–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sakiyama H, Fujiwara N, Noguchi T, Eguchi H, Yoshihara D, Uyeda K, Suzuki K. 2010. The role of O-linked GlcNAc modification on the glucose response of ChREBP. Biochem. Biophys. Res. Commun. 402:784–789 [DOI] [PubMed] [Google Scholar]
- 17. Adamson AW, Suchankova G, Rufo C, Nakamura MT, Teran-Garcia M, Clarke SD, Gettys TW. 2006. Hepatocyte nuclear factor-4alpha contributes to carbohydrate-induced transcriptional activation of hepatic fatty acid synthase. Biochem. J. 399:285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cha J, Repa JJ. 2007. The liver X receptor (LXR) and hepatic lipogenesis. The carbohydrate-response element-binding protein is a target gene of LXR. J. Biol. Chem. 282:743–751 [DOI] [PubMed] [Google Scholar]
- 19. Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. 2009. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 89:147–191 [DOI] [PubMed] [Google Scholar]
- 20. Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. 1999. Identification of a nuclear receptor for bile acids. Science 284:1362–1365 [DOI] [PubMed] [Google Scholar]
- 21. Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. 1999. Bile acids: natural ligands for an orphan nuclear receptor. Science 284:1365–1368 [DOI] [PubMed] [Google Scholar]
- 22. Wang H, Chen J, Hollister K, Sowers LC, Forman BM. 1999. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell 3:543–553 [DOI] [PubMed] [Google Scholar]
- 23. Lambert G, Amar MJA, Guo G, Brewer HBJ, Gonzalez FJ, Sinal CJ. 2003. The farnesoid X-receptor is an essential regulator of cholesterol homeostasis. J. Biol. Chem. 278:2563–2570 [DOI] [PubMed] [Google Scholar]
- 24. Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. 2000. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102:731–744 [DOI] [PubMed] [Google Scholar]
- 25. Cariou B, van Harmelen K, Duran-Sandoval D, van Dijk T, Grefhorst A, Bouchaert E, Fruchart J, Gonzalez FJ, Kuipers F, Staels B. 2005. Transient impairment of the adaptive response to fasting in FXR-deficient mice. FEBS Lett. 579:4076–4080 [DOI] [PubMed] [Google Scholar]
- 26. Cariou B, van Harmelen K, Duran-Sandoval D, van Dijk TH, Grefhorst A, Abdelkarim M, Caron S, Torpier G, Fruchart J, Gonzalez FJ, Kuipers F, Staels B. 2006. The farnesoid X receptor modulates adiposity and peripheral insulin sensitivity in mice. J. Biol. Chem. 281:11039–11049 [DOI] [PubMed] [Google Scholar]
- 27. Ma K, Saha PK, Chan L, Moore DD. 2006. Farnesoid X receptor is essential for normal glucose homeostasis. J. Clin. Invest. 116:1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prawitt J, Abdelkarim M, Stroeve JHM, Popescu I, Duez H, Velagapudi VR, Dumont J, Bouchaert E, van Dijk TH, Lucas A, Dorchies E, Daoudi M, Lestavel S, Gonzalez FJ, Oresic M, Cariou B, Kuipers F, Caron S, Staels B. 2011. Farnesoid X receptor deficiency improves glucose homeostasis in mouse models of obesity. Diabetes 60:1861–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Duran-Sandoval D, Mautino G, Martin G, Percevault F, Barbier O, Fruchart J, Kuipers F, Staels B. 2004. Glucose regulates the expression of the farnesoid X receptor in liver. Diabetes 53:890–898 [DOI] [PubMed] [Google Scholar]
- 30. Duran-Sandoval D, Cariou B, Percevault F, Hennuyer N, Grefhorst A, van Dijk TH, Gonzalez FJ, Fruchart J, Kuipers F, Staels B. 2005. The farnesoid X receptor modulates hepatic carbohydrate metabolism during the fasting-refeeding transition. J. Biol. Chem. 280:29971–29979 [DOI] [PubMed] [Google Scholar]
- 31. Samanez CH, Caron S, Briand O, Dehondt H, Duplan I, Kuipers F, Hennuyer N, Clavey V, Staels B. 2012. The human hepatocyte cell lines IHH and HepaRG: models to study glucose, lipid and lipoprotein metabolism. Arch. Physiol. Biochem. 118:102–111 [DOI] [PubMed] [Google Scholar]
- 32. Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156–159 [DOI] [PubMed] [Google Scholar]
- 33. Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C, Guguen-Guillouzo C. 2002. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. U. S. A. 99:15655–15660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schippers IJ, Moshage H, Roelofsen H, Müller M, Heymans HS, Ruiters M, Kuipers F. 1997. Immortalized human hepatocytes as a tool for the study of hepatocytic (de-)differentiation. Cell Biol. Toxicol. 13:375–386 [DOI] [PubMed] [Google Scholar]
- 35. Caron S, Verrijken A, Mertens I, Samanez CH, Mautino G, Haas JT, Duran-Sandoval D, Prawitt J, Francque S, Vallez E, Muhr-Tailleux A, Berard I, Kuipers F, Kuivenhoven JA, Biddinger SB, Taskinen M, Van Gaal L, Staels B. 2011. Transcriptional activation of apolipoprotein CIII expression by glucose may contribute to diabetic dyslipidemia. Arterioscler. Thromb. Vasc. Biol. 31:513–519 [DOI] [PubMed] [Google Scholar]
- 36. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 37. Pellicciari R, Fiorucci S, Camaioni E, Clerici C, Costantino G, Maloney PR, Morelli A, Parks DJ, Willson TM. 2002. 6alpha-ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J. Med. Chem. 45:3569–3572 [DOI] [PubMed] [Google Scholar]
- 38. Yamada K, Noguchi T. 1999. Nutrient and hormonal regulation of pyruvate kinase gene expression. Biochem. J. 337(Pt 1):1–11 [PMC free article] [PubMed] [Google Scholar]
- 39. Bergot MO, Diaz-Guerra MJ, Puzenat N, Raymondjean M, Kahn A. 1992. Cis-regulation of the L-type pyruvate kinase gene promoter by glucose, insulin and cyclic AMP. Nucleic Acids Res. 20:1871–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Burke SJ, Collier JJ, Scott DK. 2009. cAMP prevents glucose-mediated modifications of histone H3 and recruitment of the RNA polymerase II holoenzyme to the L-PK gene promoter. J. Mol. Biol. 392:578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cha-Molstad H, Saxena G, Chen J, Shalev A. 2009. Glucose-stimulated expression of Txnip is mediated by carbohydrate response element-binding protein, p300, and histone H4 acetylation in pancreatic beta cells. J. Biol. Chem. 284:16898–16905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Watson PJ, Fairall L, Schwabe JWR. 2012. Nuclear hormone receptor co-repressors: structure and function. Mol. Cell. Endocrinol. 348:440–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Perttilä J, Huaman-Samanez C, Caron S, Tanhuanpää K, Staels B, Yki-Järvinen H, Olkkonen VM. 2012. PNPLA3 is regulated by glucose in human hepatocytes, and its I148M mutant slows down triglyceride hydrolysis. Am. J. Physiol. Endocrinol. Metab. 302:E1063–E1069 [DOI] [PubMed] [Google Scholar]
- 44. Pang S, Hsieh W, Chuang C, Chao C, Weng W, Juang H. 2009. Thioredoxin-interacting protein: an oxidative stress-related gene is upregulated by glucose in human prostate carcinoma cells. J. Mol. Endocrinol. 42:205–214 [DOI] [PubMed] [Google Scholar]
- 45. Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA. 2003. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat. Genet. 34:292–296 [DOI] [PubMed] [Google Scholar]
- 46. Gonzalez FJ. 2008. Regulation of hepatocyte nuclear factor 4 alpha-mediated transcription. Drug Metab. Pharmacokinet. 23:2–7 [DOI] [PubMed] [Google Scholar]
- 47. Gineste R, Sirvent A, Paumelle R, Helleboid S, Aquilina A, Darteil R, Hum DW, Fruchart J, Staels B. 2008. Phosphorylation of farnesoid X receptor by protein kinase C promotes its transcriptional activity. Mol. Endocrinol. 22:2433–2447 [DOI] [PubMed] [Google Scholar]
- 48. Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. 2005. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature 437:759–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li M, Yang X. 2011. A retrospective on nuclear receptor regulation of inflammation: lessons from GR and PPARs. PPAR Res. 2011:742785 doi:10.1155/2011/742785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jeong Y, Kim D, Lee YS, Kim H, Han J, Im S, Chong HK, Kwon J, Cho Y, Kim WK, Osborne TF, Horton JD, Jun H, Ahn Y, Ahn S, Cha J. 2011. Integrated expression profiling and genome-wide analysis of ChREBP targets reveals the dual role for ChREBP in glucose-regulated gene expression. PLoS One 6:e22544 doi:10.1371/journal.pone.0022544 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.