Fig 5.
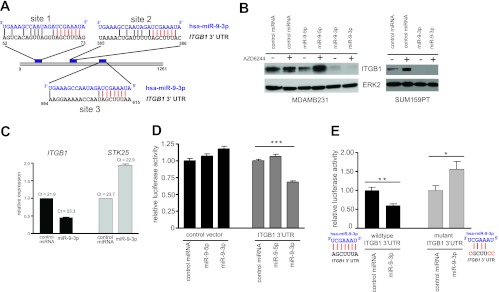
The β1 integrin gene (ITGB1) is a target of miR-9-3p. (A) Alignment of miR-9-3p with two putative binding sites (sites 1 and 2) within ITGB1 3′ UTR, as predicted by MicroCosm Targets v5 (EMBL-EBI). A third site with the identical seed sequence homology but weaker overall homology to miR-9-3p is shown as site 3. Seed sequence identity is highlighted in red. (B) (Left) Western blot showing the loss of endogenous ITGB1 protein in MDA-MB-231 cells 96 h posttransfection with the miR-9-3p mimic but not the miR-9-5p mimic in the presence or absence of 500 nM AZD6244; (right) loss of endogenous ITGB1 protein in SUM159PT cells 48 h after transfection with the miR-9-3p mimic in the presence or absence of 250 nM AZD6244. (C) Real-time PCR showing decreased levels of the ITGB1 transcript in SUM159PT cells 48 h after miR-9-3p mimic transfection. STK25 transcript levels are shown as a negative control. (D) Dual-luciferase reporter assay showing miR-9-3p dampening of the firefly luciferase transcript linked to the full-length human ITGB1 3′ UTR. The firefly luciferase signal was normalized to the signal obtained for Renilla luciferase to control for transfection efficiency, and this ratio was then normalized to the value for control miRNA-treated cells for control or ITGB1 3′ UTR vectors. Error bars indicate SEMs of 10 replicate wells within a 96-well assay plate. ***, P < 0.001. (E) Loss of miR-9-3p-mediated suppression of luciferase-ITGB1 3′ UTR reporter expression with mutation of three predicted miR-9-3p binding sites (A). *, P < 0.05, **, P < 0.01. The ITGB1 3′ UTR nucleotide substitutions designed to disrupt miR-9-3p seed sequence binding are indicated in red.
