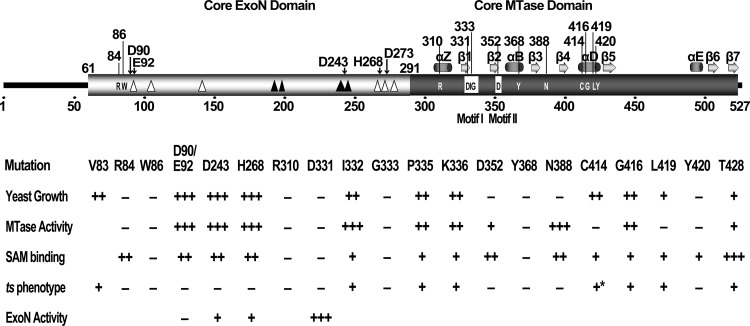
Fig 5.
Summary of the predicted domain structure of SARS-CoV nsp14 and the mutational effects of critical residues in different assays. The core domains of ExoN and MTase are indicated with open and solid black boxes, respectively. The predicted secondary structure of MTase is shown above the MTase domain, where α indicates α-helix and β represents β-sheet. The critical residues for MTase activity are shown inside the domain boxes of nsp14 (with residue positions above the boxes). The boxed residues indicate the conserved motif I (SAM-binding site) and motif II of the MTase. The open triangles indicate the internal deletions of 2 to 5 amino acid residues which did not affect the MTase activity, and the solid black triangles indicate the lethal internal deletions which inactivated the MTase. The conserved residues and positions of the DEDDh motif of ExoN are indicted by black arrows above the structure box. The arabic numbers indicate the positions or scales of the residues. The effects and phenotypes of the mutations of the critical residues are listed in the lower panel. All amino acids are mutated to alanine except H268L, C414R, and G416R. Results of five different assays are shown: yeast growth, the assays in yeast complementation system; MTase activity, in vitro biochemical assays; SAM binding, in vitro binding assay; ts phenotype, temperature-sensitive tests in yeast system (the asterisk indicates the ts phenotype identified in virus replication); and ExoN activity, in vitro exonuclease assay. The values corresponding to +++, ++, +, and − represent >60%, >20%, <20%, and < 5% of the activity of the wild type (or wild-type growth), except for the ts phenotype, in which + and − indicate the ts phenotype and lethality observed in yeast cells.