Fig 8.
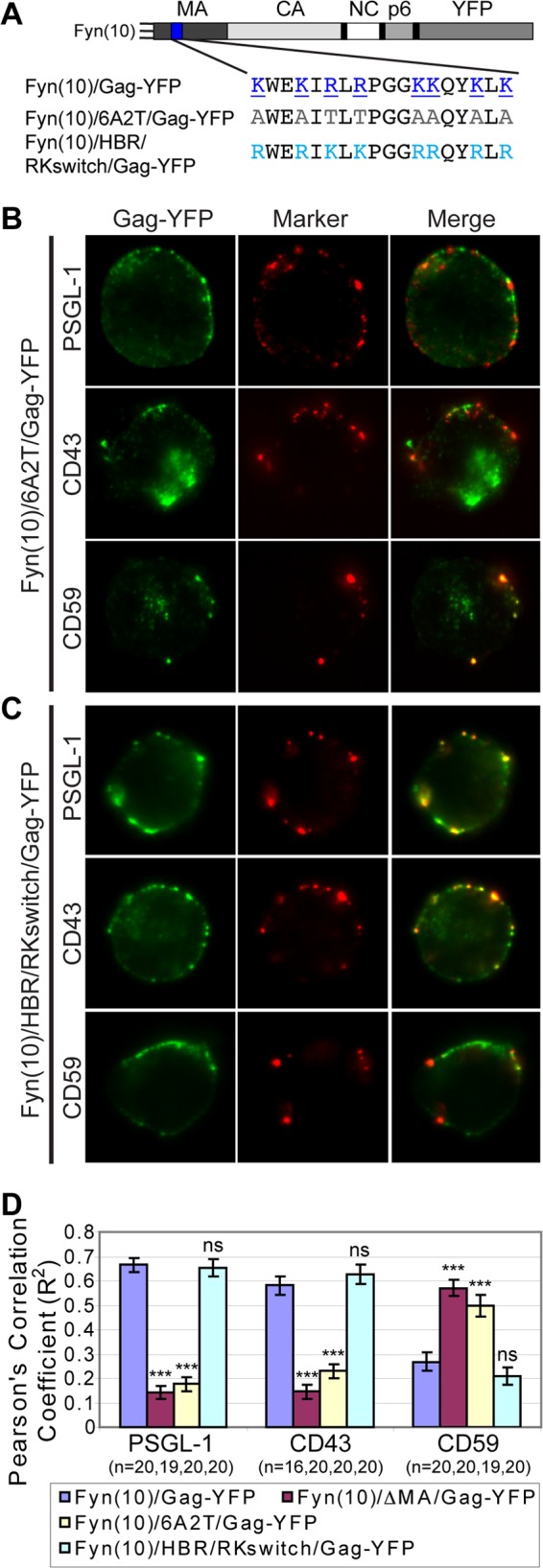
A positive charge of the MA HBR is required for specific UDM association. (A) Illustration depicting the MA HBR amino acid sequences and changes introduced in the MA mutants 6A2T and HBR/RKswitch. The basic amino acids of the wild-type HBR (blue, underlined) were changed to neutral amino acids (gray) in 6A2T or switched from R to K and from K to R (cyan) in HBR/RKswitch. (B and C) P2 cells were nucleofected with molecular clones encoding Fyn(10)/6A2T/Gag-YFP (B) or Fyn(10)/HBR/RKswitch/Gag-YFP (C). After 3 days, cells were immunostained before fixation for PSGL-1, CD43, or CD59. The middle focal plane of cells is shown. (D) Copatching between Gag and uropod markers was quantified with the square of Pearson's correlation coefficient (R2) as described for Fig. 7. n, the number of cells used for quantification. Error bars represent standard errors of the means. Data for Fyn(10)/Gag-YFP and Fyn(10)ΔMA/Gag-YFP from Fig. 7 are included for comparison. P values were calculated based on comparisons between Fyn(10)/Gag-YFP and its derivatives. ***, P < 0.001; ns, not significant.
