Fig 10.
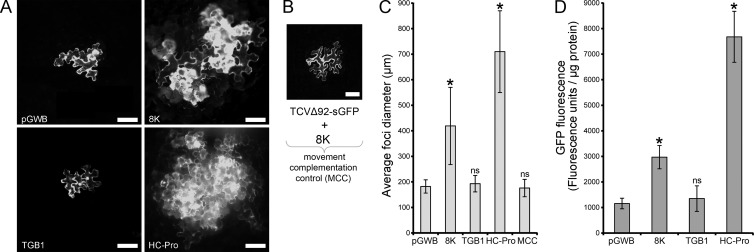
Analysis of local RNA silencing suppression activity in N. benthamiana plants. (A) Representative images of TCV-sGFP complementation assay with constructs indicated on the panels. Preinfiltrated Agrobacterium cultures carrying either an empty plasmid (pGWB) or TGB1 construct does not complement the movement of TCV-sGFP, while preinfiltrated 8K and PVA HC-Pro constructs complement the movement of TCV-sGFP at 3 dpi. Scale bar, 100 μm. (B) Representative image showing the lack of movement. Preinfiltrated Agrobacterium cultures carrying the 8K construct do not complement the movement of TCVΔ92-sGFP at 3 dpi. Scale bar, 100 μm. (C) Quantification of the movement of TCV-sGFP in the complementation assay. The diameter of 40 foci of infection were measured at 3 dpi for each construct indicated below the graph. An asterisk (*) indicates a statistically significant (P < 0.0005) difference in focus diameter compared to the empty plasmid control (pGWB). ns, not significant (P > 0.05). (C) GFP fluorometric analysis of the protein extracts from N. benthamiana leaves of the PZP-TCV-sGFP coinfiltration assay. Leaves were coinfiltrated with PZP-TCV-sGFP and various constructs, indicated below the graph. The baseline represents the level of fluorescence of the protein extract from the leaf expressing an empty T-DNA. Both PMTV 8K and PVA HC-Pro showed statistically significant increases in GFP fluorescence (P < 0.0005). ns, not significant (P > 0.05). Error bars denote standard deviation of the measurements.