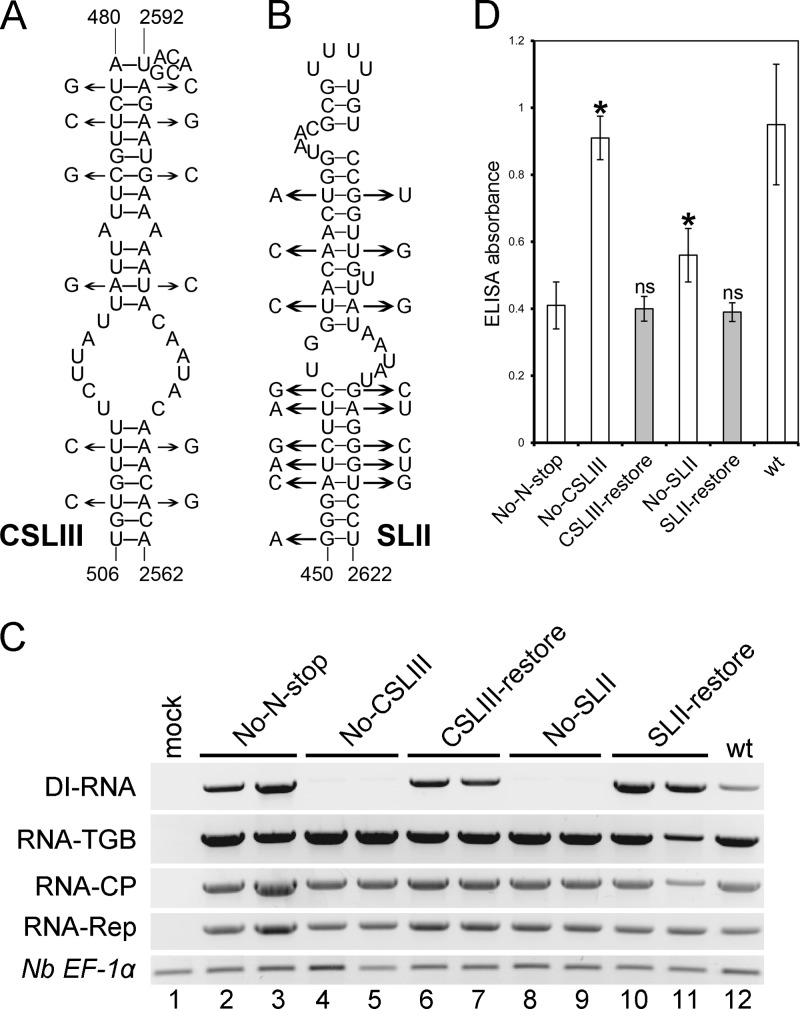
Fig 7.
Effects of CSLIII and SLII mutations on accumulation of PMTV DI RNA and the virions. (A and B) Mutations used to disrupt the stem-loops are shown on the left side of the secondary structures and refer to the No-CSLIII and No-SLII mutants, respectively. Mutations introduced into the No-CSLIII and No-SLII mutants to restore the stem-loops are shown on the right side of the secondary structures and refer to the CSLIII-restore and No-SLII-restore mutants, respectively. Numbering refers to nucleotide positions in the plus strand of full-length RNA-TGB. (C) RT-PCR of cDNA of N. benthamiana upper leaves to detect the accumulation of DI RNA and the virus genomic RNAs. The identity of each mutant is indicated above the gels. Twenty-four cycles were used for amplification. The constitutively expressed gene for N. benthamiana elongation factor 1α (Nb EF1α) served as a normalization control. The experiment was repeated twice with similar results. (D) Levels of PMTV virion accumulation detected by double-antibody sandwich ELISA as indicated by absorbance values at 405 nm. Plant extracts were prepared from upper leaves at 14 dpi. The average absorbance of a healthy plant extract was equal to 0.16. Asterisks indicate that the absorbance values were significantly different (P < 0.05, Student t test) from No-N-stop, which was used as a backbone to obtain all other constructs. ELISAs were conducted twice (n = 12).