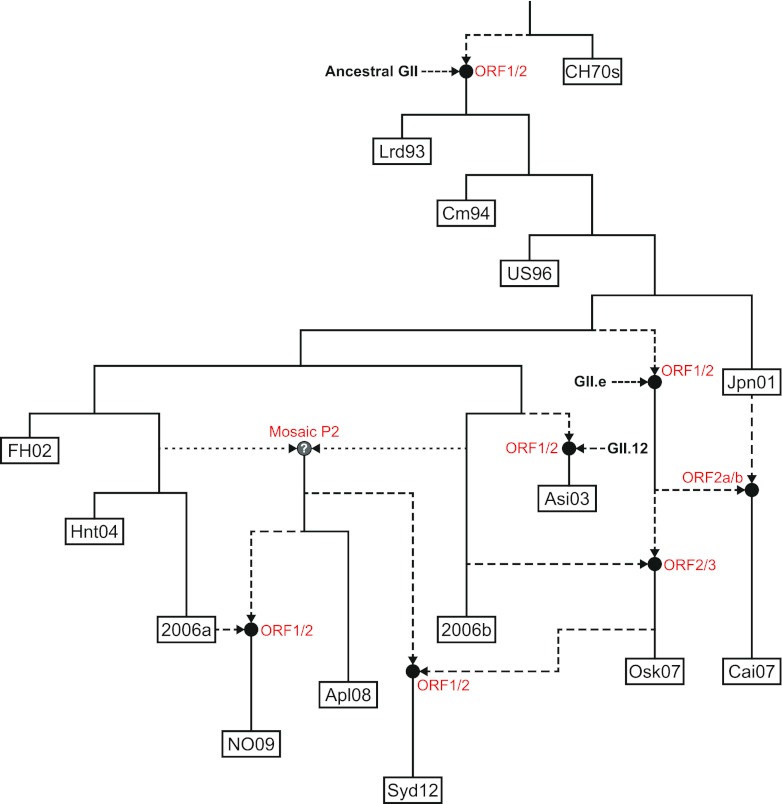
Fig 7.
Model for the emergence and origin of the NoV GII.4 variants. On the basis of evidence collected from the phylogenetic and recombination analyses, the evolution of the NoV GII.4 lineage through time was reconstructed, showing the influence of recombination. Each of the following major GII.4 variants is shown as a labeled box: CHDC 1970s (CH70s), Lordsdale 1993 (Lrd93), Camberwell 1994 (Cm94), US 1995/96 (US96), Japan 2001 (Jpn01), Farmington Hills 2002 (FH02), Asia 2003 (Asi03), Hunter 2004 (Hnt04), 2006a, 2006b, Osaka 2007 (Osk07), Cairo 2007 (Cai07), Apeldoorn 2008 (Apl08), New Orleans 2009 (NO09), and Sydney 2012 (Syd12). Black circles represent recombination events, with the dashed lines showing the direction of the nonvertical evolution from parental strains. The locations of the breakpoints are shown next to each recombination event in red text. The mosaic P2 domain recombination leading to the emergence of the Apeldoorn 2008-like viruses was not confirmed in this study; therefore, it has been shown with dotted lines and a question mark in the recombination node. Together, these results highlight the widespread recombination in the GII.4 lineage and its impact on the emergence of novel variants.