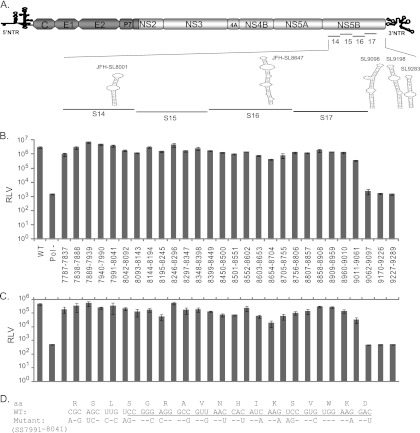
Fig 5.
High-resolution mutational analysis for identification of additional cis-acting replication elements in the NS5B protein-encoding region. (A) Schematic diagram of HCV genome depicting the mutated subsegments in NS5B. JFH-1 CRE stem-loops present in and adjacent to the mutated subsegments are presented. SL9332 and SL9389 are not shown. (B) Genome replication of mutant reporter viruses. A total of 28 contiguous mutant reporter viruses were constructed in NS5B (covering nucleotide positions 7787 to 9289), and a full-length RNA genome was generated for each mutant reporter virus by in vitro transcription. Huh-7.5.1 cells were electroporated with individual mutant RNA genomes, and 72 h posttransfection, cell lysates were harvested for Renilla luciferase assay. Means of triplicate values were calculated and are presented in the bar graph with standard deviations. The nucleotide positions of each mutant are given below the bar graph. FNX-Rluc wild-type and polymerase activity-null mutant reporter viruses were included as controls. (C) The corresponding infectivities of mutant reporter viruses presented in panel B are shown in the bar graph as mean values with standard deviations. The Renilla luciferase activities were measured from the Huh-7.5.1 cell lysates harvested 48 h postinfection. Mutants SS8195-8245, SS8654-8704, SS9011-9061, SS9062-9097, and SS9170-9226, exhibiting over 5-fold reductions (P value < 0.005) in infectivity compared to that of the wild-type virus, were selected for detailed study. (D) Nucleotide sequence of subsegment SS7991-8041. The nucleotide substitutions engineered in the mutant virus are depicted. The sequence forming the predicted stem-loop JFH-SL8001 is underlined. The screening experiment was done twice, and data from a representative experiment are shown.