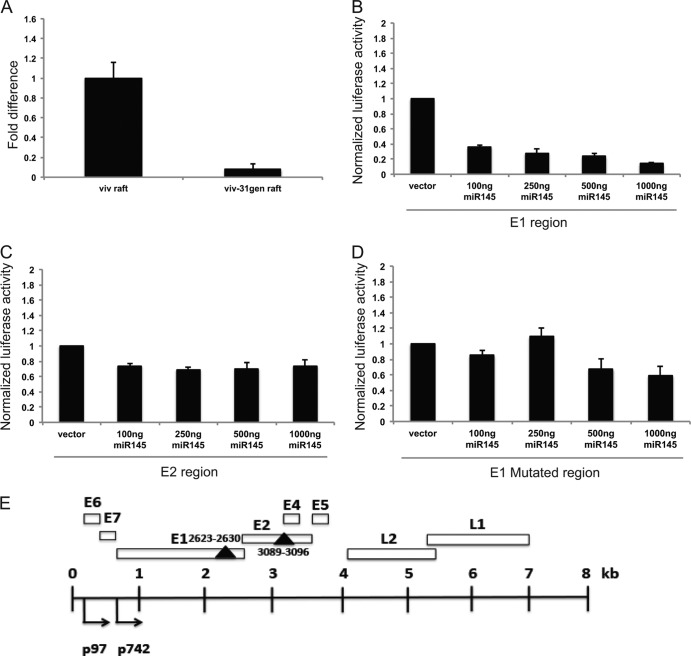
Fig 1.
Levels of miR-145 are repressed in HPV-31-positive organotypic rafts. (A) qPCR analysis of miR-145 levels in 13-day-old organotypic raft cultures of matched normal keratinocytes (viv) and HPV-positive cells that stably maintain episomes (viv-31gen). The data are normalized to a commonly used control, U6 small RNA levels, and are represented as fold changes with respect to miR-145 levels in viv raft cultures, which were set to 1.0. U6 RNA is commonly used as a normalization control for miRNA studies. (B and C) Luciferase reporter assays measuring responsiveness to miR-145 of sequences in HPV-31 E1 and E2. The two miR-145 target regions were individually cloned into a 3′UTR-luciferase reporter (psiCheck-2; Promega), and the relative luciferase activity (Renilla/firefly luciferase) of each region in the presence of increasing concentrations of miR-145 expression plasmid was assayed in transient assays. The miR-145 sequence in E1 showed a significant reduction (80%) in luciferase activity with increasing levels of miR-145 (B), whereas the E2 region showed a slight reduction in luciferase activity (C). The data are from three independent experiments, and standard errors are shown. (D) Mutation of miR-145 seed sequence in the E1 region (AAC TGG AAA converted to AAT TGG AAA) resulted in loss of responsiveness to miR-145. The data shown are averages with standard errors from three independent experiments. (E) Schematic representation of HPV-31 genome with miR-145 target sequences indicated. The early and late promoters, designated p97 and p742, are indicated along with various open reading frames, which are shown as open boxes. The two solid triangles represent the miR-145 seed sequences, and the nucleotide positions are indicated.