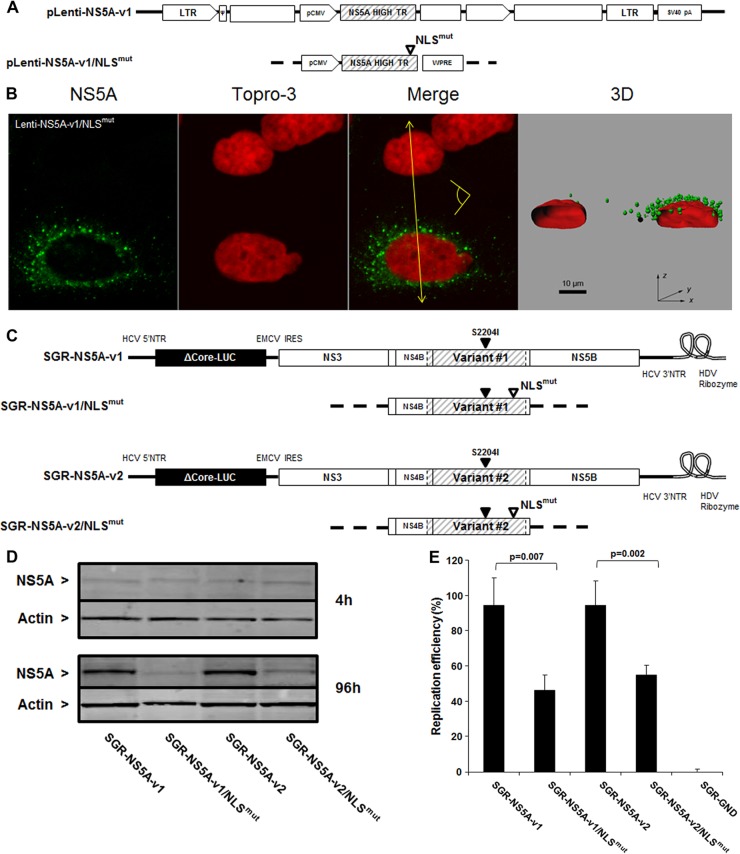
Fig 10.
Contribution of NS5A nuclear localization signal to HCV RNA replication. (A) Schematic representation of lentivirus vectors carrying mutated NS5A-v1. The open triangle denotes invalidation of the NS5A NLS. (B) Confocal analysis of cells transduced with the mutated NS5A-v1 lentiviruses was carried out as described in the legend to Fig. 4. Scale bar, 10 μm. *, nontransduced cells. The yellow arrow shows where the 3D reconstructed image was cropped to display the cell sections. The yellow pictogram indicates the direction of sight. (C) Schematic representation of subgenomic replicons encoding NS5A-v1 or -v2 containing mutated NLS. An open triangle denotes invalidation of the NS5A NLS, and a closed triangle indicates the presence of the cell culture adaptive mutation S2204I. (D) Western blotting of NS5A expression in cells transfected with NLS-mutated and nonmutated v1 and v2 replicons at 4 and 96 h posttransfection. Actin expression was used as a loading control. Bands corresponding to 56-kDa NS5A or 42-kDa actin are indicated. (E) Replication efficiency of HCV subgenomic replicons carrying NS5A variants encoding mutated NLS was analyzed in the cell lysates 96 h posttransduction. The replication efficiencies were calculated as described in the legend to Fig. 3C. The data are indicative of the mean ± SEM replication efficiencies obtained from 7 independent experiments carried out in triplicate. The data were normalized to the corresponding nonmutated controls (v1 or v2). Statistical significance was analyzed by means of a Mann-Whitney test.