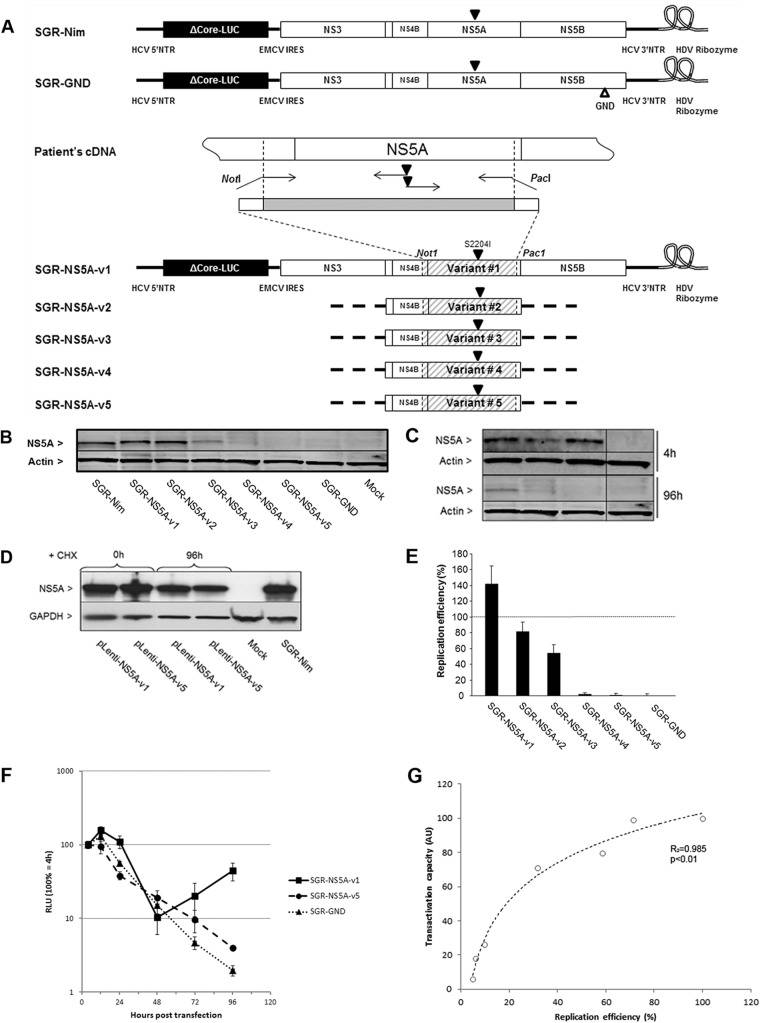
Fig 3.
Impact of HCV NS5A transactivation properties on replication efficiency of subgenomic HCV replicons harboring different NS5A quasispecies variants. (A) Schematic representation of subgenomic replicon constructs encoding NS5A-v1 to -v5. NS5A variant sequences were cloned into a subgenomic replicon shuttle vector. Solid triangles indicate the introduction of cell culture adaptive mutation S2204I. An open triangle indicates the introduction of the GDD-to-GND mutation in the NS5B RNA-dependent RNA polymerase. SGR-NS5A-vX are subgenomic replicons carrying NS5A variant sequence vX; SGR-Nim is the wt replicon (24); SGR-GND is the nonreplicative control replicon. NTR, nontranslated region; EMCV, encephalomyocarditis virus; IRES, internal ribosome entry site; HDV, hepatitis delta virus. (B) NS5A protein expression in NG cells transfected with subgenomic replicons encoding different NS5A variants as analyzed by Western blotting at 96 h posttransfection. Actin expression was used as a loading control. Bands corresponding to 56-kDa NS5A or 42-kDa actin are indicated. (C) NS5A protein expression in NG cells transfected with subgenomic replicons encoding NS5A-v3, -v4, and -v5 was analyzed by Western blotting at 4 and 96 h posttransfection. Actin expression was used as a loading control. Bands corresponding to 56-kDa NS5A or 42-kDa actin are indicated. (D) NG cells transduced with NS5A-v1 and -v5 lentiviral vectors were treated with cycloheximide (CHX) and analyzed for NS5A expression by Western blotting at 0 and 96 h posttreatment. (E) Replication efficiency of HCV subgenomic replicons carrying different NS5A variants was analyzed in the cell lysates as detailed in panel B. Luciferase activities were assayed in transfected cells, and the replication efficiency was calculated as a percentage of that obtained with replicon SGR-Nim (represented by the dashed line). The data are indicative of mean ± SEM replication efficiencies obtained from 7 independent experiments carried out in triplicate. (F) Replication efficiency of HCV subgenomic replicons carrying the v1 and v5 NS5A variants was analyzed as a time course in the cell lysates. Luciferase activities were assayed in transfected cells and are presented as a percentage of that obtained at 4 h. Data are indicative of mean ± SEM replication efficiencies obtained from 4 independent experiments. (G) Relationship between the transcriptional activation capacities of NS5A variants and the replication efficiencies of the corresponding subgenomic replicons. Data points represent the mean transactivational capacities and replication efficiencies for each variant.