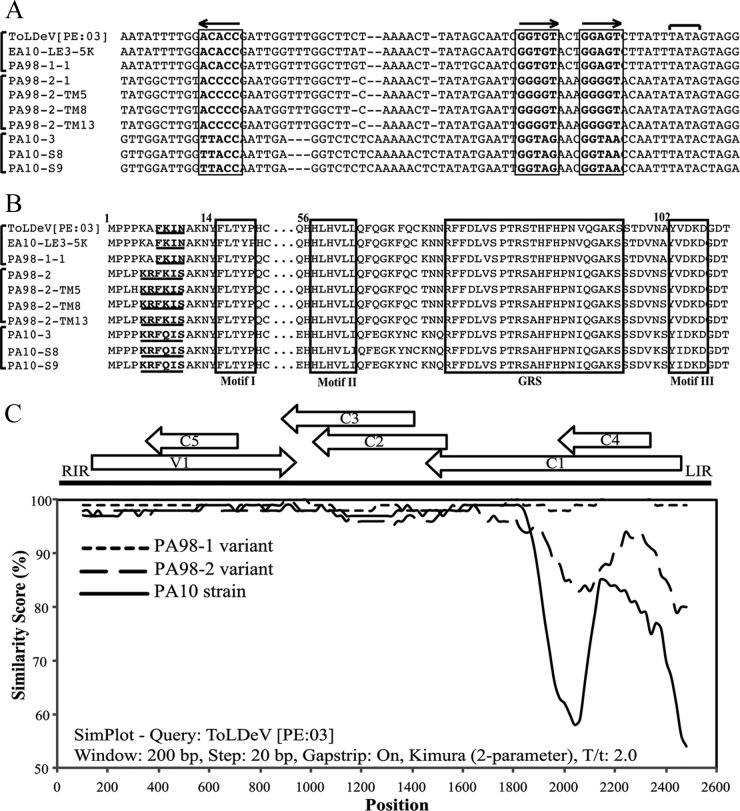
Fig 2.
Genetic diversity among ToLDeV isolates collected from Peru in 1998 (PA98-1-1, PA98-2-1, PA98-2-TM5, PA98-2-TM8, and PA98-2-TM13), 2003 (ToLDeV [PE:03]; GenBank accession number GQ335572), and 2010 (PA10-3, PA10-S8, and PA10-S9), and Ecuador in 2010 (EA10-LE3-5K). (A) An alignment of intergenic region nucleotide sequences with the Rep binding site. The sequence and orientation of the Rep binding site iteron and inverted repeat are indicated with boxes and arrows, respectively; the TATA box of the C1 (Rep) gene is indicated by a horizontal bracket. Vertical brackets show isolates with identical iteron and inverted repeat sequences and that correspond to the PA98-1 and PA98-2 variants and the PA10 strain of ToLDeV. (B) An alignment of N-terminal Rep protein amino acid sequences with the IRD. The IRD is shown in bold and underlined, and the Rep motifs I, II, and III and GRS are shown in boxes. Vertical brackets show isolates with identical IRD sequences and that correspond to the PA98-1 and PA98-2 variants and the PA10 strain of ToLDeV. (C) A similarity plot analysis of total nucleotide sequences of the genomic DNA of isolates of ToLDeV collected from Peru in 1998 and 2010 and Ecuador in 2010. The PA98-1 variant data are for isolates PA98-1-1 and EA10-LE3-5K, the PA98-2 variant data are for isolates PA98-2-1, -TM5, -TM8, and -TM13, and the PA10 strain data are for isolates PA10-3, -S8, and -S9.