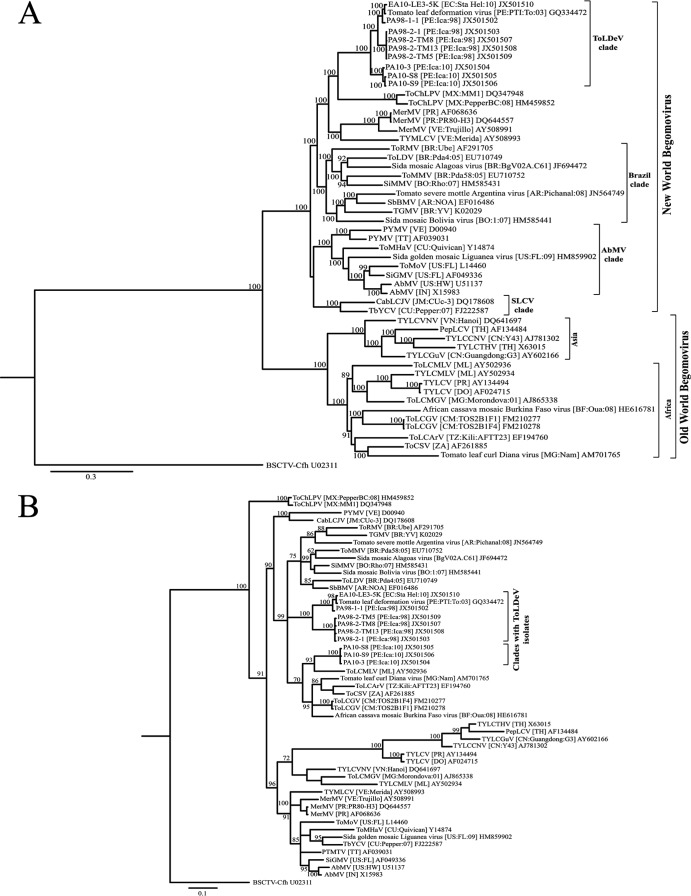
Fig 3.
Phylogenetic consensus trees showing the relationships among ToLDeV isolates collected from Peru in 1998 (PA98-1-1, PA98-2-1, PA98-2-TM5, PA98-2-TM8, and PA98-2-TM13), 2003 (ToLDeV [PE:03]; GenBank accession number GQ335572), and 2010 (PA10-3, PA10-S8, and PA10-S9) and Ecuador in 2010 (EA10-LE3-5K) and the most closely related genomes of New World (DNA-A component) and Old World begomoviruses based on alignments of total nucleotide (A) and C4 gene (B) sequences. Phylogenetic analyses were performed with MrBayes 3.2 (35). Branch strengths were evaluated by Bayesian posterior probabilities. Nodes with clade credibility values of >60% are shown. The curtovirus BSCTV was used as an outgroup. The bar indicates length of horizontal branches corresponding to the rate of substitution/nucleotide. The following sequences were obtained from GenBank and used for comparisons and phylogenetic analysis: Abutilon mosaic virus-[Hawaii] (AbMV-[US:HW]) and AbMV-India [AbMV-IN]; African cassava mosaic Burkina Faso virus (ACMVBFV-[BF:OUa:08]); Cabbage leaf curl Jamaica virus-[Jamaica] (CabLCJV [JM:Cuc-3]); Merremia mosaic virus (MerMV)-[Puerto Rico] (MerMV-[PR]), MerMV-[PR:PR80-H3] and MerMV-Venezuela (MerMV-[VE:Trujillo]); Pepper leaf curl virus-[Thailand] (PepLCV-[TH]); Potato yellow mosaic virus-Trinidad (PYMV-[TT]) and PYMV-[VE]; Sida golden mosaic Liguanea virus-[Florida] (SiGMV-[US:FL:09]) and Sida golden mosaic virus-[Florida] (SiGMV-[US:FL]); Sida micrantha mosaic virus-[Bolivia] (SiMMV-[BO:Rho:07]); Sida mosaic Alagoas virus-[Brazil] (-[BR:BgV02A.C61]); Sida mosaic Bolivia virus-[Bolivia] (-[BO:1:07]); Soybean blistering mosaic virus-[Argentina] (SbBMV-[AR:NOA]), Tobacco yellow crinkle virus-[Cuba] (TbYCV-[CU:Pepper:07]), Tomato chino La Paz virus-[Mexico] (ToChLPV [MX:MM1]) and (ToChLPV-[MX:Pepper:BC:08]); Tomato curly stunt virus-[South Africa] (ToCSV-[ZA]); Tomato golden mosaic virus-[Brazil] (TGMV-[BR:YV]); Tomato leaf curl Arusha virus-[Tanzania] (ToLCArV-[TZ:Kili:AFTT23]); Tomato leaf curl Diana virus-[Madagascar] (-[MG:Namakely]); Tomato leaf curl Ghana virus-[Cameroon] (ToLCGV-[CM: TOS2B1F1]); ToLCGV [CM: TOS2B1F4]); Tomato leaf curl Guangdong virus-[China] (TYLCGuV [CN:Guangdong]); Tomato leaf curl Madagascar virus (ToLCMGV [MG:Morondova:01]), Tomato leaf curl Mali virus-[Mali] (ToLCMLV-[ML]); Tomato leaf distortion virus-[Brazil] (ToLDV-[BR:Pda4:05]); Tomato mild mosaic virus-[Brazil] (ToMMV-[BR:Pda58:05]); Tomato mosaic Havana virus-[Cuba] (ToMHV-[CU:Quivican]); Tomato mottle virus-[Florida] (ToMoV-[US:FL]); Tomato rugose mosaic virus-[Brazil] (ToRMV-[BR:Ube]); Tomato severe mottle Argentina virus-[Argentina] (-[AR:Pichanal:08]); Tomato yellow leaf curl China virus-[China] (TYLCCNV-[CN:Y43]); Tomato yellow leaf curl Mali virus-[Mali] (TYLCMLV-[ML]); Tomato yellow leaf curl Thailand virus-[Thailand] (TYLCTHV-[TH]); Tomato yellow leaf curl Vietnam virus-[Vietnam] (TYLCVNV-[VN:Hanoi]); Tomato yellow leaf curl virus-[Dominican Republic] (TYLCV-[DO]) and TYLCV-[PR]; Tomato yellow margin leaf curl virus-[Venezuela] (TYMLCV-[VE:Merida]). The positions of the ToLDeV isolates from Ecuador and Peru are indicated by brackets.