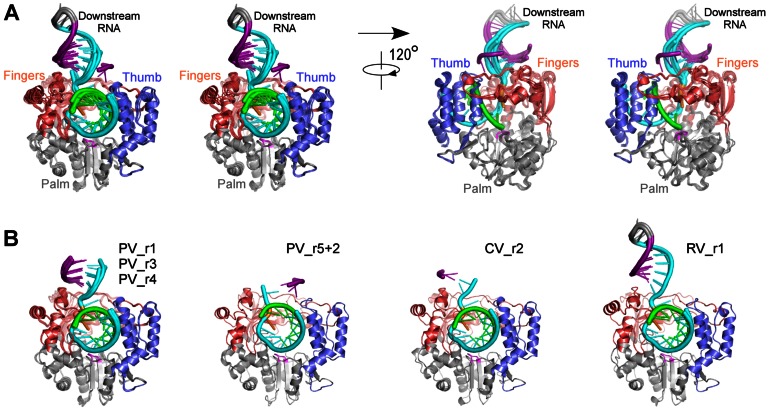
Figure 4. Comparison of EC structures and downstream RNA conformations.
A) Stereo images of all eight picornaviral EC superimposed and shown in orientations along the upstream product duplex (left) and looking into the NTP entry channel (right). The RNA is color coded as in Figure 1 and polymerases follow the three-color coding scheme with the active site YGDD residues shown in magenta. Upstream product duplex RNA nucleotides that are completely outside of the front channel (position −9 and beyond) are omitted for clarity. Note that the position of the downstream RNA duplex, protruding from the fingers domain, is consistent in all complexes except for PV_r5+2 where this RNA is rotated by almost 180° and the 5′ end is positioned above the thumb domain. B) ECs grouped according to virus species. From left to right: four PV complexes that are consistent in both polymerase and RNA conformation (two ECs from PV_r1 crystals); one PV complex that differs significantly from other PV complexes in downstream RNA conformation; one CV complex; two RV_r1 complexes that are differ slightly in downstream RNA conformation due to differences in crystal contacts (see Figure 2B).