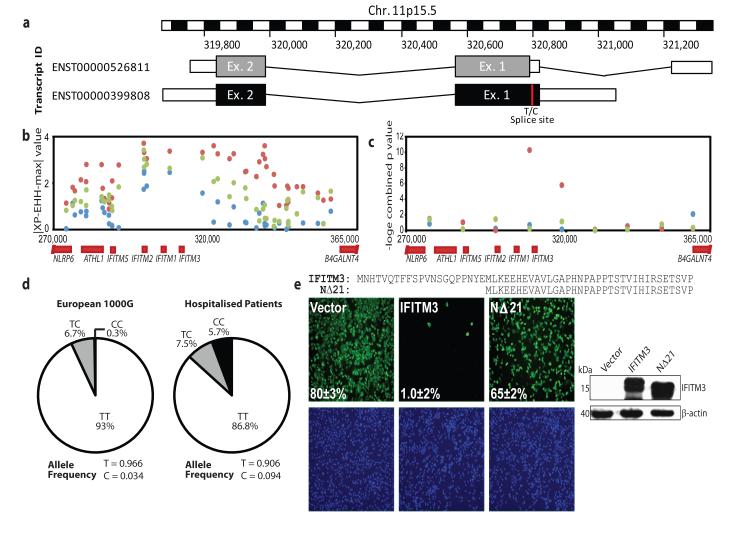
Figure 4. Single nucleotide polymorphisms of the human IFITM3 gene.
Multiple single nucleotide polymorphisms have been identified within the coding region of the human IFITM3 gene. One such SNP, rs12252 (red) (a), encodes a splice acceptor site altering T/C substitution mutation, which results in a truncated protein with an N-terminal 21 amino acid deletion. Therefore two transcripts are predicted to be expressed from the IFITM3 gene. Positive selection analysis using a haplotype-based test (∣XP-EHH-max∣, b) where data points above 2.7 in the YRI (red), 3.9 in the CEU (blue) and 5.0 in the CHB+JPT (green) are in the top 1% of values and using a combination of three allele frequency spectrum-based test statistics (c), namely Tajima’s D, Fay and Wu’s H and Nielsen et al.’s CLR on 10 kb windows along chromosome 11 encompassing the IFITM3 locus. Evidence for positive selection is seen only in the YRI. Mutations recorded through sequencing of patients hospitalised with influenza virus during the H1N1/09 pandemic showed an overrepresentation of individuals with the C allele at SNP rs12252 (d), relative to matched Europeans. A549 cells transduced to express either full-length (IFITM3) or truncated (NΔ21) IFITM3 (e) (cell nuclei: blue, virus: green, 4× magnification), show a reduction in viral restriction when IFITM3’s N-terminal 21 amino acids are removed, relative to vector controls (Vector). Alignment of the N-termini of full length (IFITM3, top) and truncated IFITM3 (NΔ21, bottom). Values represent the mean of the percent infected cells ± s.d. (n=3).