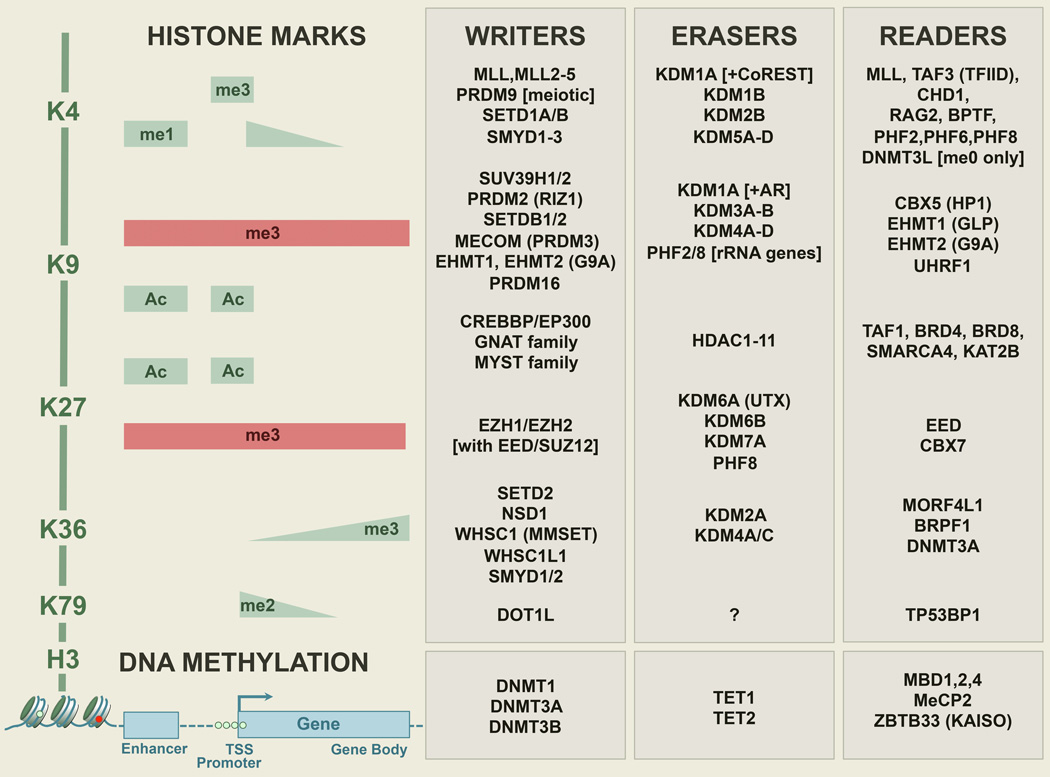
Figure 2. Histone H3 Lysine Writers, Erasers and Readers.
Although many other important histone modifications also occur, only major histone H3 lysine modifications (Ac: Acetylation; me1: monomethylation; me3: trimethylation) with well-defined functions are shown above a representative gene. The distribution of the marks are shown as colored bars and wedges to indicate approximate abundance. Repressive marks are shown in red, and active marks in blue. Epigenetic regulators are listed to the right of each mark. Acetylation across different lysines share writers and erasers, while methylation usually has dedicated enzymes. Readers (which can also be writers and erasers themselves) recognize different chromatin states and propagate the signal in various ways, including self reinforcement or cross talk, transcriptional activation or repression, or DNA repair. Crosstalk can also occur between histone modification and DNA methylation, since DNMT3A, DNMT3L, UHRF1 all contain reader domains for chromatin states.