Figure 3.
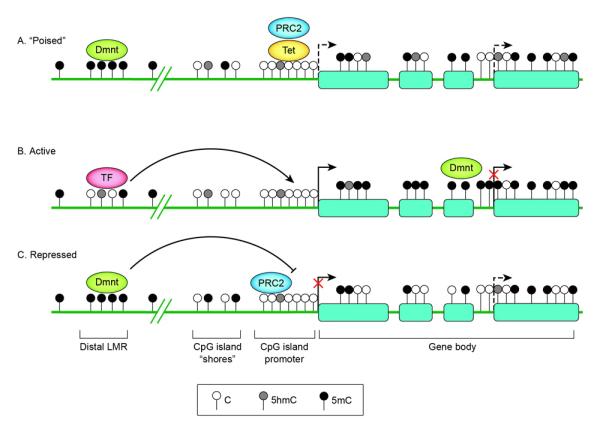
Schematic representation of DNA methylation status surrounding a gene encoding a developmental regulator in stem cells (A), in a cell displaying tissue-specific expression and differentiation towards the correct lineage (B) and in a cell differentiating towards an alternative lineage (C). At silent but “poised” promoters of genes encoding developmental regulators (A), PRC 2 mediates tri-methylation of lysine 27 of histone 3 (H3K27me3) at the promoters to keep the gene silent but “poised” for activation upon initiation of specific developmental pathways. The low-methylated regions (LMR) distal to promoters (the LMR shown here is depicted as intergenic but it can also be found at intragenic sites) form dynamically during differentiation in a cell-lineage specific manner upon binding by transcription factors (TF) [47••]. Tissue-specific DMRs have been described at CpG island (CGI) shores [37]. High-density CGI promoters generally stay unmethylated regardless of lineage fate; appropriate silencing may involve histone modifications by PRC2. Within gene bodies, methylation levels have been positively correlated with gene expression. The gene body methylation reflects a global reduction in DNA methylation observed to occur during differentiation [42,43]. Alternative TSSs are shown as dashed arrows. Full gene activation may require suppression of these alternative TSSs, possibly by intragenic CGI methylation [44•].
