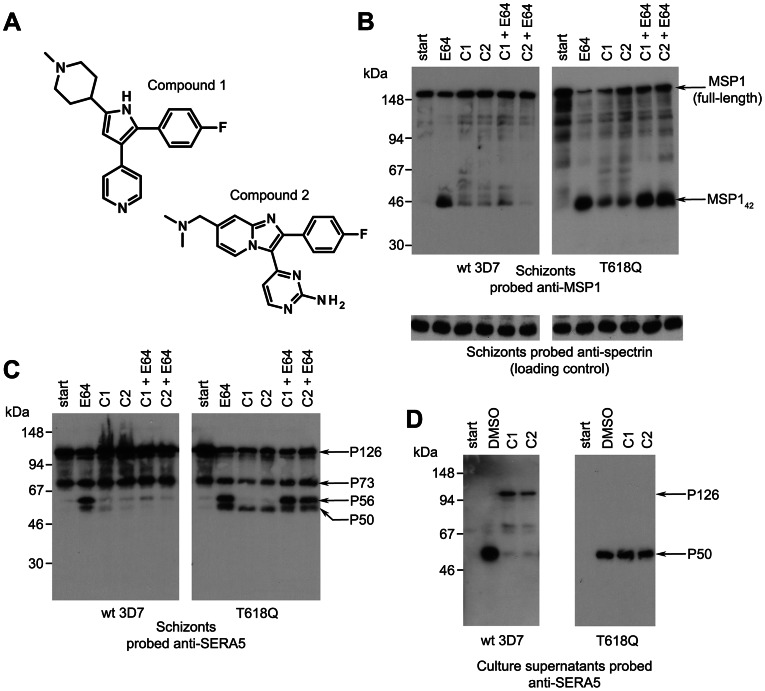
Figure 1. Structurally distinct PfPKG inhibitors block PfSUB1-mediated proteolytic processing in P. falciparum.
(A) Molecular structures of C1 and C2. (B–D) Mature wild-type (wt 3D7) or PfPKGT618Q P. falciparum schizonts were supplemented with cysteine protease inhibitor E64 only (50 µM), C1 only (2.5 µM), C2 only (1.5 µM), or combinations indicated, or vehicle only (DMSO, 1% v/v). Cultures were sampled immediately (start) or after further incubation for 4 h. SDS extracts of the residual schizonts (B, C) or culture supernatants (D) were analysed by Western blot, probing with mAbs specific for MSP1 or SERA5. An antibody to erythrocyte spectrin was used as loading control for the schizont extracts. Processing of MSP1 by PfSUB1 converts it to 4 polypeptides that include the MSP142 fragment [4]. In wt parasites, but not in the PfPKGT618Q clone, both C1 and C2 inhibited PfSUB1-mediated conversion of MSP1 to MSP142 (B) or conversion of the SERA5 precursor P126 to the processing products P56 and P50 (C), although some background processing is evidence in the presence of the inhibitors, perhaps due to imperfect synchrony of the parasite cultures. The relatively low abundance of processed SERA5 in the PfPKGT618Q schizont extracts in the presence of C1 or C2 only (C) are likely due to their loss from the schizonts at egress. Note that E64 inhibits conversion of P56 to P50, which is not mediated by PfSUB1 [1]. In (D), treatment of wt 3D7 with C1 or C2 resulted in release of only low levels of unprocessed SERA5 P126, whereas the normally-processed P50 form appeared in the PfPKGT618Q supernatants irrespective of the presence of the PfPKG inhibitors. See also Figure S1 in Text S1.