Abstract
Viral promoters are widely utilized in commercial and customized vectors to drive expression of genes of interest including reporter, effector and transfection control, because of their high transcription efficiency in a variety of primary and transformed cell lines. However, we observed altered rate of transcription for these promoters under conditions such as presence of an effector protein. These variations in viral promoter driven expressions can potentially lead to incorrect conclusion, especially in comparative and quantitative experiments. We found significantly reduced viral promoter activity in cells overexpressing tumor suppressor protein p53, whereas markedly induced transcription in cells overexpressing MAP/ERK kinase kinase 1 (Mekk 1). Using deletion constructs generated from the CMV promoter, we found the transcription reduction by p53 is possibly mediated through the TATA motif present in proximal CMV promoter. The activation of the CMV promoter by Mekk1, on the other hand, is attributed to the proximal CRE binding site in the promoter. These findings may be of interest to investigators who use CMV (or other viral) promoter driven vectors for either comparative or quantitative gene expression, or effect on promoter activity.
Keywords: Viral promoters, CMV, p53, Mekk1, JNK pathway
1. Introduction
Viral promoters are widely used as regulatory elements in mammalian expression vectors for ectopic expression of desired genes that may be reporter, effector or transfection control (e. g., β-galactosidase) protein. They show strong transcriptional activity in a variety of cells of different origin. Cytomegalovirus (CMV) promoter, for its higher transcription levels in both primary and transformed cell lines as well as in cell-free system, is the most widely used promoter in a many commercial and customized vectors. Human CMV infects a significant portion of the human population and belongs to the family of beta-herpes virus. Its promoter drives transcription of the Immediate-Early gene I of the viral genome. The core promoter of CMV has the characteristics of a typical type I promoters and contains canonical TATA box as well as initiator site in close proximity to the transcription start site. A very strong enhancer element just upstream of the core promoter contains binding sites for multiple transcription factors including NF-κB/rel, CREB/ATF, YY1, retinoic acid receptor and SP-1 (reviewed in Stinski, M. F. 1999).
p53 is an important protein in all mammlian cells and its gene is mutated in over 50% of human cancers. It prevents cell transformation by regulating cell cycle and functions as a tumor suppressor. It also serves as a transcription factor via direct DNA binding at consensus sequence located in the regulatory region of target genes and can serve as an activator or repressor depending on the target gene. The consensus sequence has internal symmetry and consists of two copies of 5'-PuPuPuC(A/T)(T/A)GpyPyPy-3’ separated by 0-13 bp.
The mammalian mitogen-activated protein kinases (MAPKs) include at least three subgroups: ERKs (extracellular signal-regulated kinases), p38 MAPKs, and JNKs (the c-Jun N-terminal kinases (Hagemann and Blank, 2001). Each is activated through a phosphorylation cascade initiated by activation of a MAP kinase kinase kinase (MAPKKK or MAP3K), which phosphorylates a MAP kinase kinase (MAPKK) that in turn phosphorylates a MAPK. Following activation, the MAPKs translocate to the nucleus to regulate the activity of transcription factors controlling a wide range of genes. One key MAP3K member, Mekk1 upon stimulation, phosphorylates Mkk4/Mkk7 (MAPKKs), which then phosphorylates and activates JNK (Maillet et al., 2009). Signaling initiated with the typically membrane-associated Mekk1 ends with activation of the transcription factor AP-1 (activator protein-1), which is a homo- or heterodimer of c-Jun with c-Fos or ATF2. Mekk1 serves mostly as an activator via AP1. However, recently we have shown that it can act as a repressor via p53 at least on polycystic kidney disease-1 gene (Islam et al, 2010).
Many laboratories utilize CMV promoter driven vectors for gene expression in experiments ranging from simply overexpressing a protein for biochemical, crystallographic or therapeutic studies to studying effects of an effector on target reporter or control genes or promoters. In comparative or quantitative experiments, investigators assume that CMV promoter itself is not affected under the experimental conditions and treatments, and therefore, does not interfere with the results. Here, we show that ectopic expression of p53 as well as of members of Mekk1-JNK-Jun/Fos signaling pathway affect CMV promoter transcription ability in transfected HEK293T cells. Furthermore, we have localized potential responsive sites for p53 and Mekk1 on the CMV promoter sequence.
2. Materials and Methods
2.1 Materials
Both pSV-β-gal control vector containing SV40 early promoter and enhancer, and pRSV-β-gal control vector with RSV promoter were purchased from Promega. pcDNA3-β-gal was a gift from Dr. Calvet lab (University of Kansas Medical Center).
The mammalian expression construct for the activated CA-Mekk1 (pFC-MEKK) was from Stratagene. This construct represents the C-terminal end (293 amino acids) of full-length (1,493 amino acids) Mekk1 (Hagemann and Blanc, 2001). p53 and dominant-negative p53 mutant were generous gifts of Dr. Samir El-Dahr (Tulane University Health Sciences Center). HCT116 p53–/– cells were a gift from the Vogelstein lab (The Johns Hopkins University).
2.2 Constructs
A 745 bp CMV promoter fragment was cut out of pCDNA3 vector using Mlu I and Xho I. This fragment contains the whole, but ~ 20 bp in the 5’, of CMV promoter, and includes the multiple cloning site upto Xho I of the vector and ligated at Mlu I-Xho I site of promoterless luciferase reporter vector pGL3-Basic (Promega). The sequential deletion constructs were created by PCR cloning using a forward primer harboring the designed Mlu I site in combination with the reverse primer GL3R1in the vector (Table 1). Following PCR, the fragment of varying sizes were restricted and cloned directionally into Mlu I-Xho I site of the pGL3 vector. The sequences were verified by sequencing.
Table 1.
Sequences of the primers used and their purposes.
| Primers | Constructs | Sequence | Purpose |
|---|---|---|---|
| GL3F | 5’-ctgctgccgaccctgtggag-3” | ChIP-qRCR | |
| GL3R1 | 5’-ggctttaccaacagtaccg-3’ | Reverse primer for PCR cloning | |
| cmv-1 | -376 | 5’-caagtgtacgcgtatgccaag-3’ | Forward primers for PCR cloning at Mlu I -Xho I site of pGL3 |
| cmv-3 | -345 | 5’-gacgtcaatgacgcgtaaatgg-3’ | |
| cmv-5 | -235 | 5’-tggtgacgcgtttttggcag-3’ | |
| cmv-7 | -125 | 5’-actaacgcgtctttccaaaatg-3’ | |
| cmv-9 | -87 | 5’-cattgacgcgtatgggcgg-3’ | |
| cmv5delf | -235del | 5’-ctccaccccatgggagtttgttttg-3’ | Deletion of CRE site in CMV-235 |
| cmv5delr | 5’-caaaacaaactcccatggggtggag-3’ |
The putative CRE binding site in H-235 was deleted using the primer pairs cmv5delf anc cmv5delr (Table 1) and the QuikChange site-directed mutagenesis kit (Stratagene) following the Supplier's instruction. The construct -43 after the TATA box was cloned using the unique Sac I site.
All PCRs were performed in similar fashion: following a denaturation step at 95°C for 3 min, 30 cycles of 94°C for 30 sec, respective annealed temperature (55-60°C) for 45 sec, and 72°C for 60 sec, with a final extension of 7 min at 72°C.
2.3 Cells, transfections and reporter assays
HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 4.5 g/liter glucose. The media was supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) and antibiotic (100 IU/ml penicillin, 100 μg/ml streptomycin). HCT116 p53–/– cells were maintained in McCoy's medium containing 10% FBS and antibiotic). All cells were grown at 37°C supplied with 5% CO2. Following overnight culture in six-well (or 12-well) plates, the cells were transfected using the calcium phosphate method. After 6 hr, the DNA-containing medium was removed, and the cells were incubated with growth media, and harvested at 40 hours as described earlier (Islam et al.,2010). In some cases, the transfected cells were treated with vehicle DMSO or JNK-specific inhibitor, SP600125 in DMSO (10 μM). Luciferase assays were carried out in 1x reporter lysis buffer (Promega) using Luminol (Promega) as a substrate. Whenever applicable, β-galactosidase assays were performed using o-nitrophenyl-β-D-galactopyranoside (Sigma) as a substrate. Protein concentrations were determined with the BCA protein assay kit (Pierce). The measured luciferase activity in each sample was normalized to protein concentration. These normalized luciferase activities (RLU) were plotted using Microsoft Excel as average ± SD of triplicate samples from typical experiments. Experiments were repeated at least three times.
2.4 SDS-PAGE and Western Immunoblotting
Expression of p53 and Cam in cells was confirmed by 10 % SDS-PAGE, followed by immunoblotting using respective primary antibodies: rabbit anti-p53 (SantaCruz, sc-6243, 1:1,500), rabbit anti-Mekk1 (SantaCruz, sc-252, 1:1,500). Alkaline phosphate-conjugated secondary anti-rabbit (Sigma, A8025) antibodies were used at 1:10,000 dilution. Following equilibration in chemiluminescent buffer (0.1 M diethanolamine, 1 mM MgCl2, pH 9.5) for 5 min, the membrane was incubated with substrate CDP-Star (Amersham) for 5 min and exposed to film (RPI).
Chromatin immunoprecipitation (ChIP)-qPCR
HEK293T cells or HCT p53-/- cells in p60 plates transfected with CMV-235, CMV235D, CMV-87 and CMV-47 were crosslinked quenched, sonicated and immunoprecipitated as described earlier (Islamt et al., 2010). Antibodies used were rabbit preimmune antisera (Sigma), rabbit anti-p53 (SantaCruz, sc-6243), or rabbit anti-cJun (SantaCruz, sc-1694). Following un-crosslinked, the DNA from the supernatants was purified using MinElute Reaction cleanup kit from Qaigen. Real-time (quantitative) PCR of the input and specific and nonspecific immunoprecipitated DNA was performed using the forward primer GL3F and the reverse primer GL3R1 given in Table 1 and GoTaq qPCR master mix (Promega) in a total reaction volume of 25 ml, and the products were detected by SYBER Green in a Bio-Rad CFX96 cycler. The products were confirmed by melting temperatures. The qPCR condition was: 95° C for 3 min and then 40 cycles of three steps: 30 sec at 95° C, 62 sec at 62° C and 45 sec at 72° C. The “cycle threshold” Ct values were selected from the linear part of the fluorescence signal between 300-400 RFU. ΔCt was determined as: ΔCt = Avg. PKD1 Ct – Avg. L7 Ct, and gene-fold expression was calculated according to the Livak method, 2-ΔΔCt. The fluorescent signals from the specific and nonspecific immunoprecipitated DNA was normalized with that of the respective input DNA. These normalized fluorescence units (RLU) were plotted using Microsoft Excel as average ± SD of triplicate.
3. Results and Discussion
3.1 Viral promoters are affected by ectopically expressed p53 and CAM
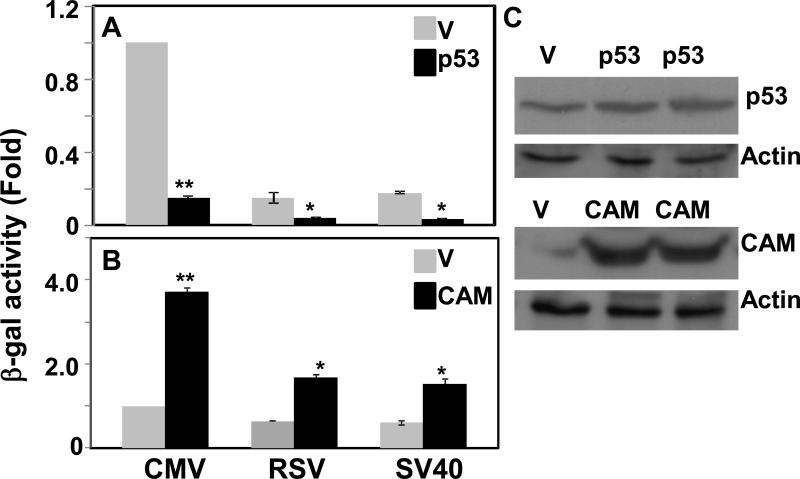
Viral promoters, especially CMV, RSV and SV40, are widely used in commercial and customized vectors for expression of target genes for various purposes including characterization, biochemical studies or even therapeutic use of the protein products. They are also used extensively to express effector proteins such as transcription factors to investigate latter's effects, or to express reporter genes such as β-galactosidase, β-glucuronidase, or luciferase as a normalization control for quantitative gene expression or promoter activities. In these and similar experiments, investigators naturally assume that the reporter control gene expressing vectors are not subject to change with various experimental condition applied. While working with β-galactosidase expression vector as a transfection control in our experiments, we unexpectedly noticed its expression driven by CMV promoter was drastically affected with ectopically expressed p53 or constitutively active Mekk1 (CAM) (Fig. 1) compared to co-transfected empty pcDNA3 vector in HEK29T cells. Whether vectors containing other viral promoters such as RSV and SV40 are similarly affected, we examined the effects of overexpressed p53 or CAM on the expression of β-galactosidase driven by either RSV (pRSV-gal) or SV40 (pSV-gal) in transiently transfected HEK293T cells. As shown in Fig. 1, both RSV and SV40 promoters showed similar trend in effects as observed with the CMV promoter by overexpressed p53 and CAM. These results indicated that viral promoters currently used widely in expression vectors are not independent of and subject to change to the experimental conditions. Such changes in expression in comparative and/or quantitative experiments of the target gene, reporter, control or promoter may lead to erroneous conclusion.
Fig.1.
Effect of ectopically expressed p53 (A) and CAM (Constitutively Active Mekk1, B) on expressions of β–galactosidase from vectors with CMV (pcDNA3-gal), RSV (pRSV-gal) or SV40 (pSV-gal) promoters. 293T cells were transfected with indicated promoter (CMV, RSV or SV40) containing vectors with empty vector (V), p53 cDNA or CAM cDNA in pcDNA3. β-galactosidase was assayed in cell lysates after 48 hours. All β-galactosidase values were normalized to protein concentration and are shown as means standard deviations (*, p < 0.01; **, p < 0.005; p53 or CAM versus V). The normalized β-galactosidase activity in cells transfected with pcDNA3-gal and empty pcDNA3 vector was set to 1.0. Data of mean were obtained from at least three different experiments. (C) immunoblots of lysates of HEK293T cells transfected with control, CAM or p53 expression vectors probed with anti-Mekk1 or anti-p53 antibody. β-actin (Actin) was used as a loading control.
3.2 CMV promoter repression by p53 is mediated by -87 bp proximal, core promoter sequence
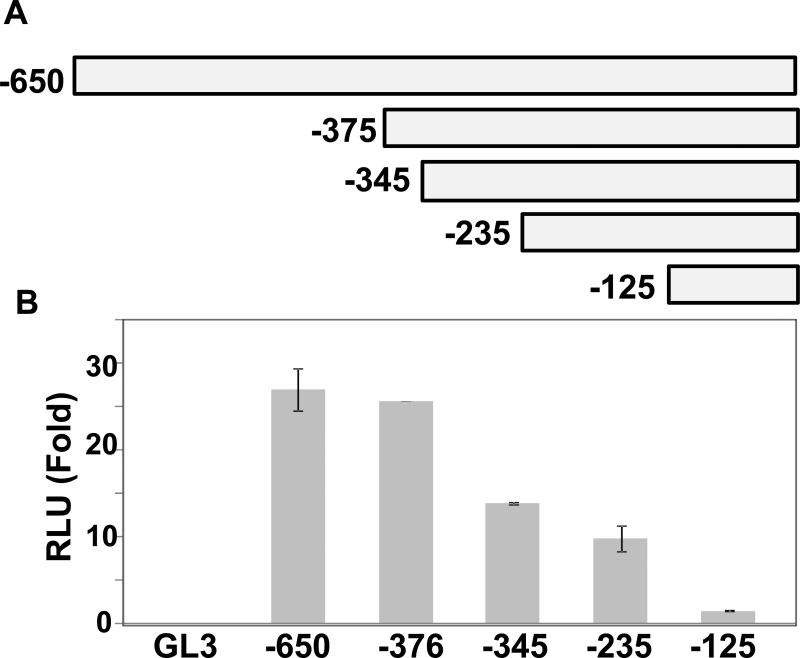
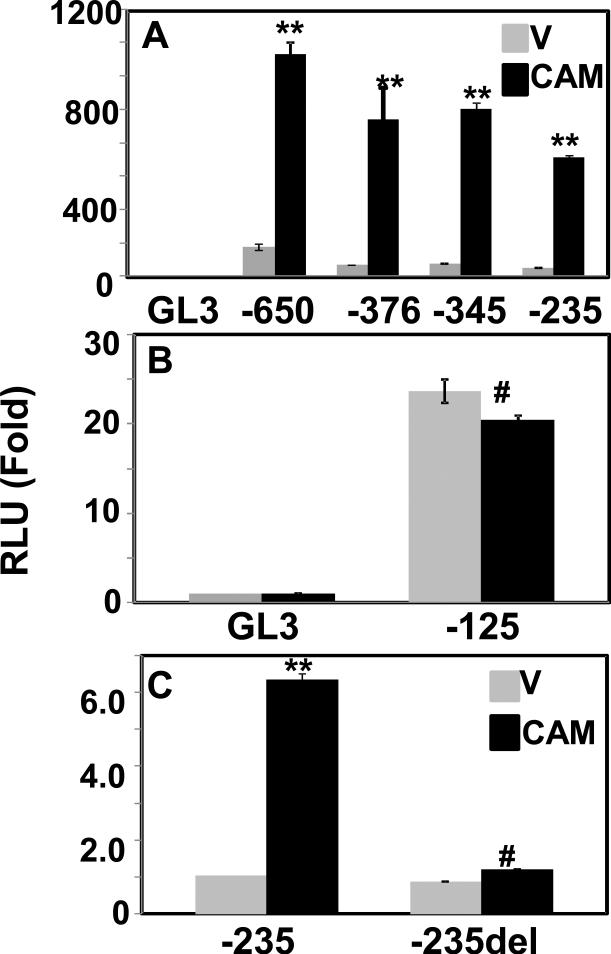
To gain further insight, we focused on CMV promoter (Fig. 2) as it is widely utilized in expression vectors. We excised the 650 bp CMV promoter fragment from pcDNA3 vector using Mlu I and Xho I enzymes and inserted into pGL3 basic vector containing a luciferase gene downstream as reporter, and then made a series of serial deletions in the promoter fragment (Fig.3A). Following that, we examined the intrinsic promoter activity of each of these deleted promoter in pGL3 vector in HEK293T cells. As shown in Fig. 3B, interestingly the CMV-375 construct had promoter activity, measured as luciferase output, comparable to the full length (-650) CMV promoter, suggesting the promoter fragment between -650 and -370 is not essential, at least in HEK293T cells, for gene expression or promoter function. Further shortening of the promoter progressively reduced expression of the luciferase reporter in these cells. This was expected as deletion might have eliminated binding sites for enhancer and/or other cellular factors.
Fig. 2.
The CMV promoter sequence. The transcription initiation site is assigned as position +1, and indicated by arrow. The nucleotide positions upstream are presented as negative numbers. The start of each deleted construct is shown with a slash and underlined number. The potential CRE consensus binding site and TATA motif are shown in bold-underlined.
Fig.3.
Intrinsic promoter activity of serially deleted CMV promoter constructs. (A) Schematic representation of deletion constructs generated from CMV promoter fused to luciferase reporter gene in pGL3-Basic vector. The numbers indicate nucleotide positions in the CMV promoter in 5’of the transcription start site. (B) HEK293T cells were transfected with constructs shown in A or promoterless pGL3-Basic (GL3), and luciferase activity was measured in cell extracts after 48 h of transfection. The luciferase activity in each transfectant was normalized to β-galactosidase expressed in pcDNA3, and is shown as means standard deviations. The normalized RLU in cells transfected with pGL3-Basic was set to 1.0. Data of mean were obtained from at least three different experiments.
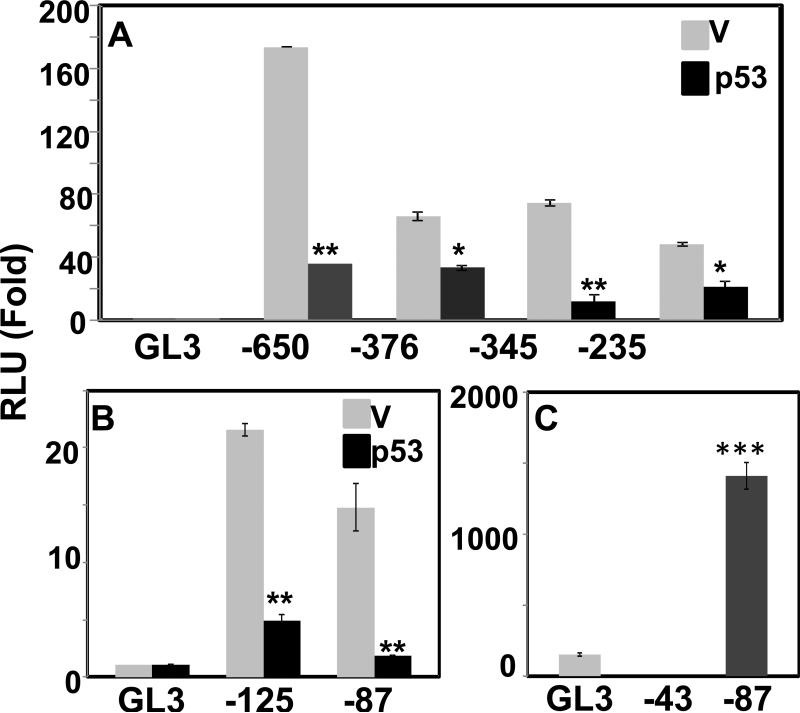
Next, we examined effects of ectopic expression of p53 on promoter activity of all the deleted constructs in HEK293T cells. The results in Fig.4A-B illustrated all the truncated constructs including CMV-87 were repressed by p53 similar to the full-length CMV promoter. The extent of repression observed in CMV-87 was comparable to the full length CMV-650, indicating a sequence in this short, proximal fragment harbors a p53 responsive site. Upon inspection of CMV-87 region we were unable to find any p53 consensus site, but identified a TATA box. TATA box is a core promoter element that binds TBP (TATA binding protein), which is an indispensable member of basic transcriptional machinery in all TATA containing promoters. Mutations or deletion of TATA box disrupts TBP binding leading to loss of any transcriptional activity. This was evident in our experiment (Fig. 4C) with CMV-43 without the TATA box, which lost drastically (2000 times vs CMV-87) transcriptional activity to the point that it caused significant suppression of the background activity of pGL3 basic, most likely by interfering with transcription machinery.
Fig.4.
Effect of overexpressed p53 on the deleted CMV promoter constructs. (A-B) HEK293T cells were co-transfected with pGL3-Basic vector (GL3) or deleted CMV promoter constructs in pGL3 together with pcDNA3 vector (V) or p53 in pcDNA3 vector (p53). Luciferase activity was measured at 48 h after transfection. The luciferase activity in each transfectant was normalized as in Fig. 1 and is shown as means standard deviations (*, p < 0.01; **, p < 0.005; p53 versus V). The normalized RLU in cells co-transfected with pGL3-Basic and pcDNA3 was set to 1.0. Data of mean were obtained from at least three different experiments. (C) Intrinsic promoter activity of CMV promoter constructs -87 containing TATA box and -43 without TATA box. HEK293T cells were co-transfected with pGL3-Basic vector (GL3), CMV-87 or CMV-43 promoter constructs in pGL3 together with β–galactosidase expression plasmid. The luciferase activity in each transfectant was normalized to β-galactosidase expressed, and is shown as means standard deviations. The normalized RLU in cells transfected with pGL3-Basic was set to 1.0. Data of mean were obtained from at least three different experiments (***, p < 0.001; versus GL3).
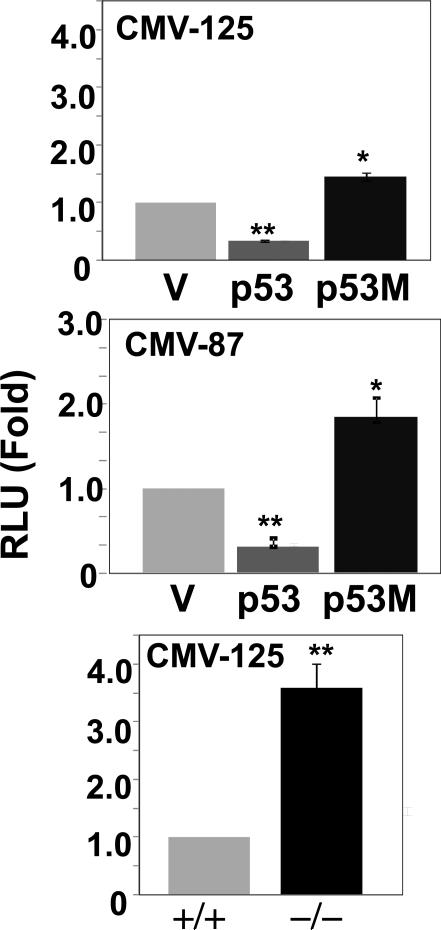
To demonstrate the repression effect of p53 on the CMV promoter is specific, we investigated activities of the two shortest promoter constructs, CMV-125 and CMV-87, co-transfected with expression plasmids for either wild-type p53 or mutated p53 (p53M). This p53M harbors a mutation in its DNA binding domain abolishing its binding capability to the consensus site. As shown in Fig.5 A-B, promoter activities of both constructs in HEK293T cells co-transfected with p53M were comparable or even slightly higher than those in cells co-transfected with the empty pcDNA3, whereas their activities were significantly reduced in cells co-transfected with the wild type p53. To provide further support, next we measured intrinsic promoter activity of the CMV-125 construct in HCT116 cells with genetically ablated p53 (p53 -/-). In comparison to promoter activity in the wild type cells (p53+/+) more than 3-fold increased activity was noticed in cells with ablated p53 (Fig. 5C). Taken together, these and earlier results demonstrated that p53 specifically represses CMV promoter through a site located in the 87 bp of proximal promoter.
Fig. 5.
Effect of mutated p53 and ablated p53 on CMV-125 and CMV-87 promoter constructs. CMV-125 or CMV-87 construct was co-transfected into HEK293T cells along with wild type p53 (p53), mutated p53 (p53M, a DNA binding site mutant) or their expression vector, pcDNA3 as control (V). Luciferase activity was determined at 48 h after transfection. The luciferase activity in each transfectant was normalized as in Fig. 1 and is shown as means standard deviations (*, p < 0.01; **, p < 0.005; p53 or p53M versus V). The normalized RLU in cells transfected with CMV-125 or CMV-87 together with control pcDNA3 vector was set to 1.0. The intrinsic promoter activity of CMV-125 construct was measured after 48 h of transfection into HCT116 cells (+/+) or HCT 116 cells lacking p53 (-/-). The normalized RLU in p53 (+/+) cells was set to 1.0. The results are presented as RLU fold change (**, p < 0.005; -/- versus +/+).
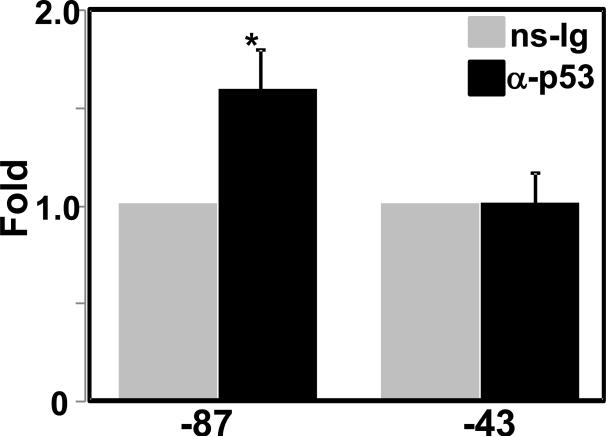
The transcription factor p53 is a well-known tumor suppressor which is able to activate gene transcription by binding to specific palindromic sequences (el-Deiry et al., 1992). The consensus p53 binding site consists of two half-sites, each comprising two copies of the sequence PuPuPuC(A/T) arranged head-to-head and separated by 0–13 nucleotides (Levine, 1997). On the other hand, there may be alternative modes of p53 binding to target DNA sequences that significantly deviate from the canonical consensus in terms of sequence and/or spatial arrangement. p53 has also been shown to repress transcription of a number of genes (Venot et al., 1998). Genes repressed by p53 can be divided into two general types (Johnson et al., 2001). In most promoters of the first type, p53 exerts its repression not by direct DNA binding but rather through an interaction with a promoter-bound transcriptional activator such as Sp1 (Kanaya et al., 2000), NF-Y (Yun et al., 1999), C/EBPbeta (Kubicka et al., 1999) and AP-1 (Sun et al., 1999). Repression of the second type of promoters occurs through the minimal promoter region, and p53 decreases promoter activity of these genes by interaction with basal transcriptional machinery. It has been shown that p53 interacts directly with the TATA-binding factor, TBP (Truant et al., 1993), as well as with the TBP-associated factors dTAFII40 and dTAFII60 (Farmer et al., 1996) and other basal factors (Thut et al., 1995). TBP bound to p53 is unable to associate with other proteins of the basal transcription machinery (Ragimov et al., 1993; Crighton et al., 2003). Suppression by p53 of the minimal promoter containing only initiator element and TATA motif has been shown to depend on p53 binding with TBP (Seto et al., 1992; Mack et al., 1993; Ragimov et al., 1993). p53 mediated transcriptional suppression of insulin-like growth factor I receptor gene promoter also involves p53 interaction with TBP (Werner et al., 1996). Similar mechanism of repression by p53 has been reported for Bcl-2 and Cox-2 promoters (Wu et al., 2001; Subbaramaiah et al., 1999). In these promoters, progressive deletions up to the minimal region containing TATA box maintained repression by p53, suggesting that TATA sequence in the basal promoter is responsible for the p53 mediated repression. The proximal CMV-87 promoter construct was still repressed in cells with co-expressed p53. This minimal promoter does not contain p53 consensus binding site, but contains a TATA-motif. As mentioned earlier, TBP is indispensable member of basic transcriptional machinery of TATA containing promoters and disruption of TBP binding leads to loss of transcriptional activity. As such the CMV-43 construct without the TATA box drastically lost transcriptional activity- 2000 times lowered than TATA containing CMV-87 construct (Fig. 4C). Thus, we were unable to demonstrate involvement of TATA motif in p53 mediated repression of the CMV promoter using deletion method. However, ChIP-qPCR analysis (Fig. 6) showed significantly more p53 binding with CMV-87 promoter containing the TATA motif compared to CMV-43 construct without TATA box, suggesting interaction of p53 with the TATA motif. Taken together, these results on the CMV promoter provide another example of p53 repression by interference with TATA motif and basal transcription machinery.
Fig. 6.
Association of p53 with the CMV promoter was evaluated using ChIP–qPCR analysis. Chromatin DNA form HEK293T cells transfected with CMV-83 or CMV-47 were cross-linked, fragmented, and immunoprecipitated using anti-p53 (α-p53) or preimmune (ns-Ig) sera prior to real-time PCR as described in the method. The amplified values in each were normalized with respective input amplified value, and normalized ns-Ig value for each transfected sample was set to 1.0. The bar is a mean (*, p < 0.01; α-p53 versus ns-Ig).
3.3 CMV promoter activation is mediated by JNK-AP1 pathway and CRE site
To dissect the CAM activation effect on expression vectors driven by viral promoters (Fig. 1), again we focused on the CMV promoter and used the serially deleted promoter-luciferase constructs created in Fig. 3A. In transiently co-transfection experiments, we observed that all but CMV-125 constructs were strongly activated by co-expressed CAM (Fig. 7A-B). Inspecting the promoter region between -235 and -125, we identified a consensus cyclic AMP responsive element (CRE) site at -166 (Fig. 2). CRE sites are found in promoters regulated by cAMP. In these promoters, ubiquitous transcription factor CRE binding protein (CREB) binds DNA at consensus CRE site, TGACGTCA (Montminy and Bilezikjian, 1987). Well documented downstream targets of activated Mekk1 are JNK, a MAPK, which in turn activates transcription factor, AP-1 (Activated Protein-1). AP-1 is a homo/hetero dimer of Jun with Fos or ATF (Shaulian and Karin, 2001) and activates transcription of target genes through binding at AP-1 consensus DNA sequence, 5’-TGAG/CTCA (Angel et al., 1987). Comparison of AP-1 and CREB consensus DNA binding sites, a similar DNA sequence emerged and there is evidence that AP-1 and CREB family proteins able to dimerize and bind to CRE sequences, allowing cross-talk between the two signaling pathways (Benbrook and Jones, 1990). To examine this possibility, we created CMV-235del. Deletion of the CRE sequence in the construct (CMV-235del) abolished its ability to be activated by co-transfected CAM as illustrated in Fig. 7C. These results indicated that the CRE binding site at -166 in the CMV promoter might serve as the responsible site for CAM mediated activation.
Fig.7.
Effect of ectopically expressed constitutively active Mekk1 (CAM) on transcriptional activity of serially deleted CMV promoter constructs. (A-B) HEK293T cells were co-transfected with indicated CMV deletion reporter constructs (shown in Fig.3A) or empty pGL3-Basic (GL3) together with empty vector pcDNA3 (V) or CAM expression plasmid in pcDNA3 (CAM). Luciferase activity was assayed 48 h after transfection. Transfection efficiencies were normalized as in Fig. 1 and expressed as RLU means SD (*, p < 0.01; **, p < 0.005; CAM versus V). The normalized RLU in pGL3-Basic with control pcDNA3 transfected cells was set to 1.0. (C) Similar to (A) but transfected with CMV-235 and CMV-235del (a derivative of CMV-235 lacking CRE binding site at -166 position).The normalized RLU in CMV-235 with pcDNA3 control vector transfected cells was set to 1.0 (**, p < 0.005; #, not significant; CAM versus V)
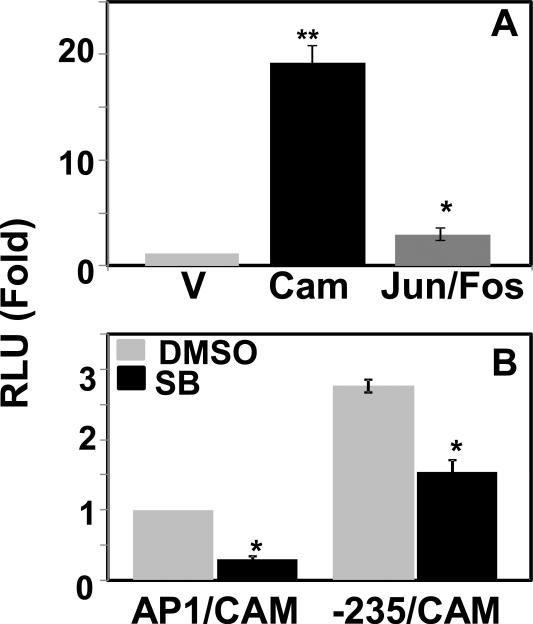
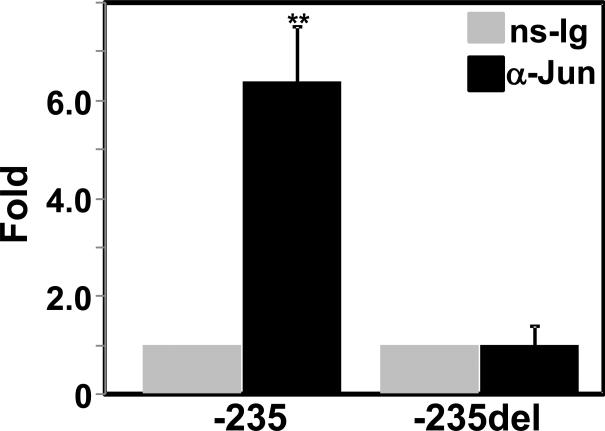
As Mekk1 activates AP-1 transcription factor via activation of JNK, we investigated whether AP-1 and JNK are involved in the Mekk1 mediated activation of the CMV promoter. AP-1 is a homo-or heterodimeric protein composed of members of c-Jun with c-Fos or ATF protein families (Shaulian and Karin, 2001).The construct CMV-235 was also activated when cotransfected with the members of AP-1, Jun and Fos expression plasmids (Fig. 8A). Activation of the CMV-235 construct and a JNK pathway control promoter, AP1 (which contains five tandem consensus sites for AP1) by CAM was significantly reduced in the presence of JNK specific inhibitor SB600125 (Fig. 8B) suggesting involvement of Mekk1-JNK-AP1 pathway. To obtain further support that AP-1 is involved, ChIP-qPCR analysis (Fig. 9) demonstrated ~8-fold more binding with the CMV-235 promoter compared to CMV-235del when immunoprecipitated using anti-Jun antibody. Taken together, these data suggest that the JNK/AP-1 pathway is involved in CAM mediated activation of the CMV promoter through the CRE site at -166.
Fig. 8.
JNK-pathway is involved in CAM mediated activation of the CMV promoter. (A) CMV-235 promoter reporter construct was transfected into HEK293T cells along with empty pcDNA3 vector, CAM or Jun and Fos in pcDNA3 expression plasmids. Jun dimerizes with Fos to form transcriptional factor AP-1. Transfections were normalized and expressed as RLU means SD (*, p < 0.01; **, p < 0.005; CAM or Jun/Fos versus V). The normalized RLU in pGL3-Basic with control pcDNA3 transfected cells was set to 1.0. (B) HEK293T cells were co-transfected with AP-1 (a control promoter fragment with five consensus AP-1 binding sites in tandem) or CMV-235 construct together with CAM expression plasmid in the presence of vehicle DMSO or JNK-specific inhibitor SB600125 (10 μM) for 24 h. JNK is a MAPK downstream target of Mekk1 and immediate upstream of AP1. The luciferase activity was assayed at 48 h after transfection. Transfection efficiencies were normalized and set to 1.0 in vector transfected or DMSO treated cells, and expressed as RLU means SD (*, p < 0.01; SB600125 versus DMSO).
Fig. 9.
Association of c-Jun with the CMV promoter was evaluated using ChIP–qPCR analysis. Chromatin DNA form HEK293T cells transfected with CMV-235 or CMV-235del were cross-linked, fragmented, and immunoprecipitated using anti-Jun (α-Jun) or preimmune (ns-Ig) sera prior to real-time PCR as described in the method. The amplified values in each were normalized with respective input amplified value, and normalized ns-Ig value for each transfected sample was set to 1.0. The bar is a mean (**, p < 0.005; α-Jun versus ns-Ig).
4. Summary and conclusion
In this study, we have shown that CMV promoter activity is repressed by p53, and the TATA containing core region of the promoter is responsible for this effect. The repression is most likely mediated via interaction of p53 with TATA binding protein and/or basal transcriptional machinery on the promoter. In addition, we have shown activation of the CMV promoter by members of Mekk1-JNK-AP-1 pathway through a CRE binding site at -166 position of the promoter. Our data may be helpful to researchers in choosing appropriate vectors in reporter assays or other applications of CMV promoter containing plasmids. In conclusion, our results raise awareness that certain conditions may affect CMV and other viral promoters in expression vectors and may lead to erroneous conclusion.
Highlights-CMV.
Transcription of CMV promoter is altered by ectopic expression of p53 and Mekk1
Repression by p53 is mediated via interaction with TATA box in CMV promoter
Induction by Mekk1 is mediated through JNK/Ap1 pathway and CRE site in CMV promoter
Findings are helpful in selecting control vector in comparative gene expressions
Acknowledgement
This work was supported by NIH grants R15 DK069897 (M.R.I). We thank Dr. Vogelstein (John Hopkins University Medical Center) for HCT116 p53 null cells.
Abbreviations
- CMV
cytomegalovirus
- SV
Simian virus
- RSV
Rous Sarcoma Virus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angel P, Imagawa M, Chiu R, Stein B, Imbra RJ, Rahmsdorf HJ, Jonat C, Herrlich P, Karin M. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987;49:729–39. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbrook DM, Jones NC. Heterodimer formation between CREB and JUN proteins. Oncogene. 1990;5:295–302. [PubMed] [Google Scholar]
- Crighton D, Woiwode A, Zhang C, Mandavia N, Morton JP, Warnock LJ, Milner J, White RJ, Johnson DL. p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. EMBO J. 2003;22:2810–20. doi: 10.1093/emboj/cdg265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–9. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- Farmer G, Colgan J, Nakatani Y, Manley JL, Prives C. Functional interaction between p53, the TATA-binding protein (TBP), and TBP-associated factors in vivo. Mol. Cell. Biol. 1996;16:4295–304. doi: 10.1128/mcb.16.8.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann C, Blank JL. The ups and downs of MEK kinase interactions. Cell Signal. 2001;13:863–75. doi: 10.1016/s0898-6568(01)00220-0. [DOI] [PubMed] [Google Scholar]
- Ho JS, Ma W, Mao DY, Benchimol S. p53-Dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol. Cell. Biol. 2005;27:423–31. doi: 10.1128/MCB.25.17.7423-7431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RA, Ince TA, Scotto KW. Transcriptional repression by p53 through direct binding to a novel DNA element. J. Biol. Chem. 2001;276:27716–20. doi: 10.1074/jbc.C100121200. [DOI] [PubMed] [Google Scholar]
- Islam MR, Jimenez T, Pelham C, Rodova M, Puri S, Magenhaimer BS, Maser RL, Widmann C, Calvet JP. MAP/ERK kinase kinase 1 (MEKK1) mediates transcriptional repression by interacting with polycystic kidney disease-1 (PKD1) promoter-bound p53 tumor suppressor protein. J. Biol. Chem. 2010;285:38818–31. doi: 10.1074/jbc.M110.145284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya T, Kyo S, Hamada K, Takakura M, Kitagawa Y, Harada H, Inoue M. Adenoviral expression of p53 represses telomerase activity through down-regulation of human telomerase reverse transcriptase transcription. Clin. Cancer. Res. 2000;6:1239–47. [PubMed] [Google Scholar]
- Kubicka S, Kuhnel F, Zender L, Rudolph KL, Plumpe J, Manns M, Trautwein C. p53 represses CAAT enhancer-binding protein (C/EBP)-dependent transcription of the albumin gene. A molecular mechanism involved in viral liver infection with implications for hepatocarcinogenesis. J. Biol. Chem. 1999;274:32137–44. doi: 10.1074/jbc.274.45.32137. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–31. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Mack DH, Vartikar J, Pipas JM, Laimins LA. Specific repression of TATA-mediated but not initiator-mediated transcription by wild-type p53. Nature. 1993;363:281–3. doi: 10.1038/363281a0. [DOI] [PubMed] [Google Scholar]
- Maillet M, Lynch JM, Sanna B, York AJ, Zheng Y, Molkentin JD. Cdc42 is an antihypertrophic molecular switch in the mouse heart. J. Clin. Invest. 2009;119:3079–88. doi: 10.1172/JCI37694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montminy MR, Bilezikjian LM. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328:175–8. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- Murphy M, Hinman A, Levine AJ. Wild-type p53 negatively regulates the expression of a microtubule-associated protein. Genes Dev. 1996;10:2971–80. doi: 10.1101/gad.10.23.2971. [DOI] [PubMed] [Google Scholar]
- Ragimov N, Krauskopf A, Navot N, Rotter V, Oren M, Aloni Y. Wild-type but not mutant p53 can repress transcription initiation in vitro by interfering with the binding of basal transcription factors to the TATA motif. Oncogene. 1993;8:1183–93. [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- Seto E, Usheva A, Zambetti GP, Momand J, Horikoshi N, Weinmann R, Levine AJ, Shenk T. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc Natl Acad Sci USA. 1992;89:12028–32. doi: 10.1073/pnas.89.24.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Reddy PG, Goberdhan A, Walsh C, Dao S, Ngai I, Chou TC, O-Charoenrat P, Levine AJ, Rao PH, Stoffel A. p53 regulates cell survival by inhibiting PIK3CA in squamous cell carcinomas. Genes Dev. 2002;16:984–93. doi: 10.1101/gad.973602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinski MF. Cytomegalovirus promoter for expression in mammalian cells. In: Fernandez JM, Hoeffler JP, editors. Gene expression systems: using nature for the art of expression. Academiv Press; San Diego, Calif: 1999. pp. 211–233. [Google Scholar]
- Suubaramaiah K, Altorkil N, Chung WJ, Mestre JR, Sampat A, Dannenberg AJ. Inhibition of cyclooxygenase-2 gene expression by p53. J. Biol. Chem. 1999;274:10911–10915. doi: 10.1074/jbc.274.16.10911. [DOI] [PubMed] [Google Scholar]
- Sun Y, Zeng XR, Wenger L, Firestein GS, Cheung HS. P53 down-regulates matrix metalloproteinase-1 by targeting the communications between AP-1 and the basal transcription complex. J. Cell. Biochem. 2004;92:258–69. doi: 10.1002/jcb.20044. [DOI] [PubMed] [Google Scholar]
- Thut CJ, Chen JL, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–4. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- Truant R, Xiao H, Ingles CJ, Greenbladt J. Direct interaction between the transcriptional activation domain of human p53 and the TATA box-binding protein. J Biol Chem. 1993;268:2284–7. [PubMed] [Google Scholar]
- Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J. 1998;17:4668–79. doi: 10.1093/emboj/17.16.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner H, Karnieli E, Rauscher FJ, LeRoith D. Wild-type and mutant p53 differentially regulate transcription of the insulin-like growth factor I receptor gene. Proc Natl Acad Sci U S A. 1996;93:8318–23. doi: 10.1073/pnas.93.16.8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Mehew JW, Heckman CA, Arcinas M, Boxer LM. Negative regulation of bcl-2 expression by p53 in hematopoietic cells. Oncogene. 2001;20:240–51. doi: 10.1038/sj.onc.1204067. [DOI] [PubMed] [Google Scholar]
- Yun J, Chae HD, Choy HE, Chung J, Yoo HS, Han MH, Shin DY. p53 negatively regulates cdc2 transcription via the CCAAT-binding NF-Y transcription factor. J. Biol. Chem. 1999;274:29677–82. doi: 10.1074/jbc.274.42.29677. [DOI] [PubMed] [Google Scholar]