Fig. 5.
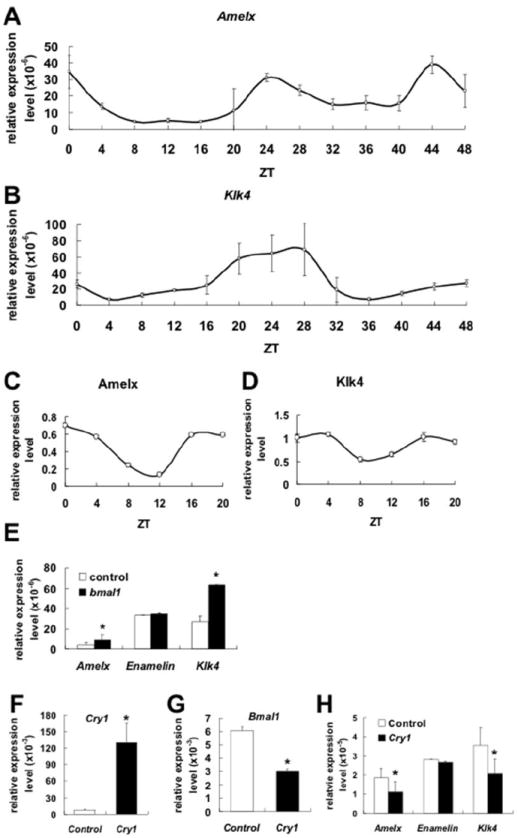
Ameloblast-specific genes expression exhibits circadian rhythms in HAT-7. A) Real-time qRT-PCR reveals that Amelx, a marker of secretory amelobalsts, RNA expression follows a circadian pattern in HAT-7 cells after cell cycle synchronization. B) Klk4, a marker of maturation ameloblsts, also exhibits RNA circadian pattern of expression. At the protein levels, after ImageJ analysis, AMELX shows the highest protein expression levels at ZT0 (C), and KLK4 protein shows the highest level of expression at ZT4 (D). Klk4 relative expression levels are slightly higher than Amelx relative expression levels in HAT-7 cells but both genes follow clear circadian patterns. Over-expression of Bmal1 cDNA results in up-regulation of Amexl and Klk4 RNAs expression in HAT-7 (E). In contrast, Enamelin (which is another ameloblast-specific gene) RNA expression remains unchanged. Over-expression of Cry1 (F) results in down-regulation of Bmal1 (G). Amelx and Klk4 expression levels also are significantly down-regulated (H). The representative data are presented as means ± S.D. from at least three independent replicates where data from cells transfected with Bmla1 or Cry1 were compared with cells transfected with empty vector. Statistical significance (*) was considered when p<0.05.
