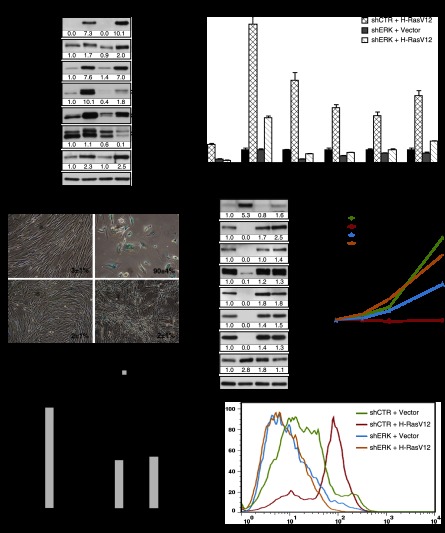
Figure 1.
ERK/MAPK inhibition bypasses Ras-induced senescence. (A) Immunoblots for proteins in the ERK pathway using extracts from fibroblasts expressing H-RasV12 (R) or an empty vector (V) and shRNA against ERK2 (shERK) or a nontargeting shRNA (shCTR) obtained from cells 14 d after infection. (B) Quantitative PCR (qPCR) for ERK2 mRNA and mRNAs encoded by ERK-stimulated genes in cells as in A. (C) SA-β-Gal of cells as in A. Data were quantified from 100 cell counts in triplicate and are presented as the mean percentage of positive cells ± standard deviation (SD). (D) Immunoblots for cell cycle-regulated proteins in cells as in A. (E) Growth curves started with cells as in A. Data are presented as mean ± SD of triplicates. (F) Quantitation of BrdU incorporation (2 h of incubation with 10 μM BrdU) and KI-67 staining in cells as in A. Data were quantified from 100 cell counts in triplicate and are presented as the mean of positive cells ± SD. (***) P < 0.0005, two-sample t-test. (G) Superoxide levels in cells as before, measured by flow cytometry (FACS) after staining with 1 μM fluorescent probe dihydroethidium (DHE) during 1 h. The results are expressed as the percentage of maximum cell number. The maximum cell number is the number of cells for the most-represented fluorescence intensity in the cell population of a condition and is expressed as 100%. All experiments were performed a minimum of three times.