Figure 5.
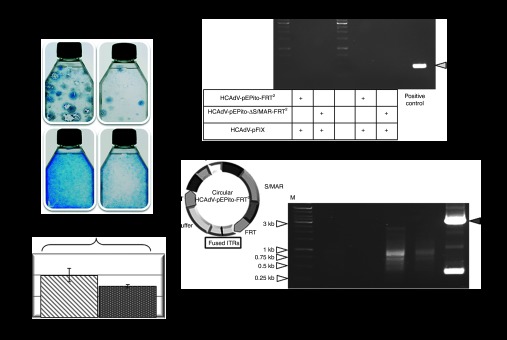
Evaluation of HCAdVs without FLPe recombination. (a) Colony forming assay performed in U87 cells to analyze maintenance of the transgene during selection pressure without FLPe recombination. Cells were infected with viruses HCAdV-pEPito-FRT2 or HCAdV-pEPito-ΔS/MAR-FRT2 at MOIs of 10 and 100. After 6 weeks under blasticidin selection pressure cell colonies were stained with methylene blue. (b) Flow cytometry 72 hours after infection with the adenoviruses HCAdV-pEPito-FRT2 or HCAdV-pEPito-ΔS/MAR-FRT2 at MOI of 50 with stuffer virus HCAdV-hFIX at MOI of 50 (total MOI of 100). The percentage of U87-expressing eGFP cells is shown. Mean values (±SD) of two independent experiments are presented. (c) Recombination PCR. U87 cells were infected at MOI of 100 for each virus. PCR was performed with primer pair Recomb-fw and Recomb-rev on genomic DNA isolated 72 hours after infection and 6 weeks after continuous blasticidin selection. As shown for the positive control a 530 bp PCR-band was expected. (d) PCR excluding ITR-fused HCAdV circles. U87 cells were either infected with HCAdV-pEPito-FRT2 and HCAdV-pEPito-ΔS/MAR-FRT2 or coinfected with the FLPe-encoding virus most likely excluding ITR-fused circles. The possible structure of HCAdV-pEPito-FRT2-genome circularized via ITR-linkage is shown in the left panel. To address the question whether giant circles of the complete adenoviral genome are formed, the primer ITR-rev binding to the ITR sequence was used. The expected PCR product spanning the potentially covalently linked adenoviral genome at the ITR sequences was 144 bp in length. The positive control shows PCR results of a PCR reaction in which the ITRs were linked by a plasmid backbone. The expected PCR product of the positive control was 3,048 bp in length (gray arrow). ITR, inverted terminal repeat; M, DNA ladder as marker; MOI, multiplicity of infections.
