SUMMARY
DNA damage elicits a cellular signaling response that initiates cell cycle arrest and DNA repair. Here we find that DNA damage triggers a critical block in glutamine metabolism, which is required for proper DNA damage responses. This block requires the mitochondrial SIRT4, which is induced by numerous genotoxic agents and represses the metabolism of glutamine into TCA cycle. SIRT4 loss leads to both increased glutamine-dependent proliferation and stress-induced genomic instability, resulting in tumorigenic phenotypes. Moreover, SIRT4 knockout mice spontaneously develop lung tumors. Our data uncover SIRT4 as an important component of the DNA damage response pathway that orchestrates a metabolic block in glutamine metabolism, cell cycle arrest and tumor suppression.
INTRODUCTION
DNA damage initiates a tightly coordinated signaling response to maintain genomic integrity by promoting cell cycle arrest and DNA repair. Upon DNA damage, ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and RAD3-related protein (ATR) are activated and induce phosphorylation of Chk1, Chk2 and γ-H2AX to trigger cell cycle arrest and to initiate assembly of DNA damage repair machinery (Abraham, 2001; Ciccia and Elledge, 2010; Su, 2006). Cell cycle arrest is a critical outcome of the DNA damage response (DDR) and defects in the DDR often lead to increased incorporation of mutations into newly synthesized DNA, the accumulation of chromosomal instability and tumor development (Abbas and Dutta, 2009; Deng, 2006; Negrini et al., 2010).
The cellular metabolic response to DNA damage is not well elucidated. Recently, it has been shown that DNA damage causes cells to upregulate the pentose phosphate pathway (PPP) to generate nucleotide precursors needed for DNA repair (Cosentino et al., 2011). Intriguingly, a related metabolic switch to increase anabolic glucose metabolism has been observed for tumor cells and is an important component of rapid generation of biomass for cell growth and proliferation (Jones and Thompson, 2009; Koppenol et al., 2011). Hence, cells exposed to genotoxic stress face a metabolic challenge; they must be able to upregulate nucleotide biosynthesis to facilitate DNA repair, while at the same time limiting proliferation and inducing cell cycle arrest to limit the accumulation of damaged DNA. The molecular events that regulate this specific metabolic program in response to DNA damage are still unclear.
Sirtuins are a highly conserved family of NAD+-dependent deacetylases, deacylases, and ADP-ribosyltransferases that play various roles in metabolism, stress response and longevity (Finkel et al., 2009; Haigis and Guarente, 2006). In this study, we studied the role of SIRT4, a mitochondria-localized sirtuin, in cellular metabolic response to DNA damage and tumorigenesis.
RESULTS
DNA damage represses glutamine metabolism
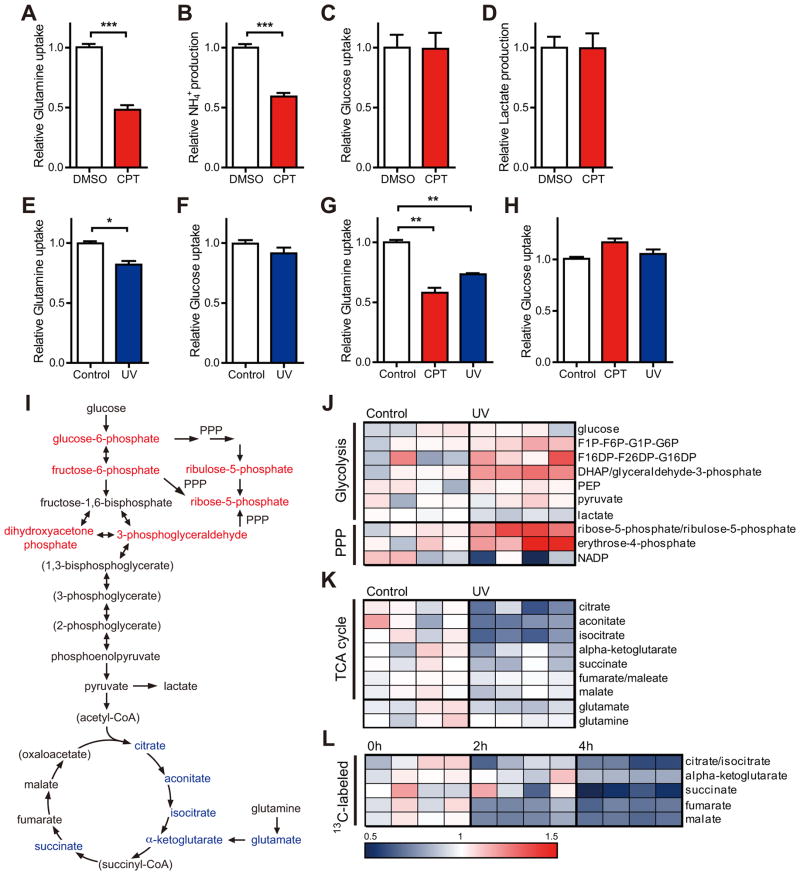
To investigate how cells might balance needs for continued nucleotide synthesis, while also preparing for cell cycle arrest, we assessed the metabolic response to DNA damage by monitoring changes in the cellular consumption of two important fuels, glucose and glutamine, after DNA-damage. Strikingly, treatment of primary mouse embryonic fibroblasts (MEFs) with camptothecin (CPT), a topoisomerase 1 inhibitor that causes double-stranded DNA breaks (DSBs), resulted in a pronounced reduction in glutamine consumption (Figure 1A). Glutamine metabolism in mammalian cells is complex and contributes to a number of metabolic pathways. Glutamine is the primary nitrogen donor for protein and nucleotide synthesis, which are essential for cell proliferation (Wise and Thompson, 2010). Additionally, glutamine provides mitochondrial anaplerosis. Glutamine can be metabolized via glutaminase (GLS) to glutamate and NH4+, and further converted to the tricarboxylic acid (TCA) cycle intermediate α-ketoglutarate via glutamate dehydrogenase (GDH) or aminotransferases. This metabolism of glutamine provides an important entry point of carbon to fuel the TCA cycle (Jones and Thompson, 2009), and accounts for the majority of ammonia production in cells (Yang et al., 2009). CPT-induced reduction of glutamine consumption was accompanied by a reduction in ammonia secretion from cells (Figure 1B). Notably, under these conditions, we observed no obvious decrease in glucose uptake and lactate production (Figures 1C and 1D), consistent with previous studies showing that intact glucose utilization through the PPP is important for a normal DNA damage response (Cosentino et al., 2011). Preservation of glucose uptake also suggests that repression of glutamine consumption may be a specific metabolic response to genotoxic stress and not reflective of a non-specific metabolic crisis.
Figure 1. Glutamine metabolism is repressed by genotoxic stress.
(A and B) Glutamine uptake (A) and ammonia production (B) in primary MEFs incubated without or with 14 μM CPT for 12 hr (n = 8–9).
(C and D) Glucose uptake (C) and lactate production (D) in primary MEFs treated as indicated in Figure. 1A (n = 8–9).
(E and F) Glutamine (E) and glucose (F) uptake in primary MEFs measured at 6 hr after 30 J/m2 UV treatment (n = 3).
(G and H) Glutamine (G) and glucose (H) uptake in HepG2 cells treated without or with 14 μM CPT for 12 hr or at 6 hr after 30 J/m2 UV treatment (n = 3–6).
(I) Schematic illustrating the metabolites that are increased (red) or decreased (blue) in transformed MEFs at 4 hr after 20 J/m2 UV exposure (n = 4; p < 0.05). Metabolites in parentheses were not measured.
(J and K) Heat map comparing relative levels of intermediates in transformed MEFs measured at 4 hr after 20 J/m2 UV treatment when compared with an untreated control (n = 4 samples of each condition). Blue and red indicates down- or upregulation, respectively.
(L) Heat map of 13C-glutamine contributed to labeled TCA cycle intermediates at 0, 2 and 4 hr after 20 J/m2 UV treatment (n = 4 samples of each condition). All changes are relative incorporation compared to each non-UV treated control after pulse of 13C-glutamine. Data are means ±SEM. *p < 0.05, **p < 0.005, ***p < 0.0001.
See also Figure S1.
To examine the metabolic response to other forms of genotoxic stress, we monitored the metabolic response to ultra-violet (UV) exposure in primary MEFs. Similar to CPT treatment, UV exposure reduced glutamine uptake, without significant changes in glucose consumption (Figures 1E and 1F). Similarly two human cell lines, HepG2 and HEK293T, also demonstrated marked reductions in glutamine uptake in response to DNA damaging agents without comparable changes in glucose uptake (Figures 1G and 1H; Figures S1A and S1B). Taken together, these results suggest that a variety of primary and tumor cell lines (from mouse or human) respond to genotoxic stress by down-regulating glutamine metabolism.
To examine in more detail the changes in cellular glutamine metabolism after genotoxic stress, we performed a global metabolomic analysis with transformed MEFs before and after DNA damage. As previously reported, we observed that PPP intermediates were increased in response to DNA damage (Figures 1I and 1J). Remarkably, we observed a decrease in measured TCA cycle intermediates after UV exposure (Figures 1I and 1K). Moreover, we found that HepG2 cells showed a similar metabolomic shift in response to DNA damage (Figure S1D). We did not observe a clear, coordinated repression of nucleotides or glutamine-derived amino acids after exposure to DNA damage (Figure S1C).
To determine whether reduction in TCA cycle metabolites was the consequence of reduced glutamine metabolism, we performed a time-course tracer study to monitor the incorporation of [U-13C5]glutamine into TCA cycle intermediates at 0, 2 and 4 hr after UV treatment. We observed that after UV exposure, cells reduced contribution of glutamine to TCA cycle intermediates in a time-dependent manner (Figure 1L). Moreover, the vast majority of the labeled fumarate and malate contained four carbon atoms derived from [U-13 C5]glutamine (Figure S1F, M+3 versus M+4), indicating that most glutamine was used in the non-reductive direction towards succinate, fumarate and malate production. We were able to observe little contribution of glutamine flux into nucleotides or glutathione in control or UV-treated cells at these time points (data not shown), suggesting that the mitochondrial metabolism of glutamine accounts for the majority of glutamine consumption in these cells. Taken together, the metabolic flux analysis demonstrates that DNA damage results in a reduction of mitochondrial glutamine anaplerosis, thus limiting the critical refueling of carbons into the TCA cycle.
To assess the functional relevance of decreased glutamine metabolism after DNA damage, we deprived cells of glucose, thereby shifting cellular dependence to glutamine to maintain viability (Choo et al., 2011; Dang, 2010). If DNA damage represses glutamine usage, we reasoned that cells would be more sensitive to glucose deprivation. Indeed, following 72 hr of glucose deprivation, cell death in primary MEFs was significantly elevated at 10 hr after UV exposure (Figure S1E). However, cells cultured with glucose remained viable in these conditions. Thus, these data demonstrate that genotoxic stress limits glutamine entry into the central mitochondrial metabolism of the TCA cycle.
SIRT4 is induced in response to genotoxic stress
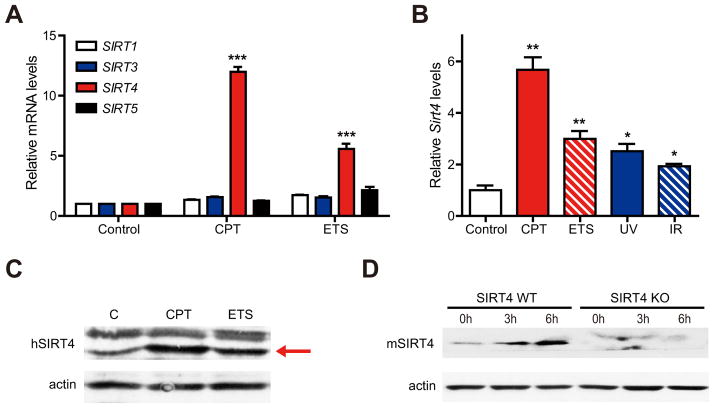
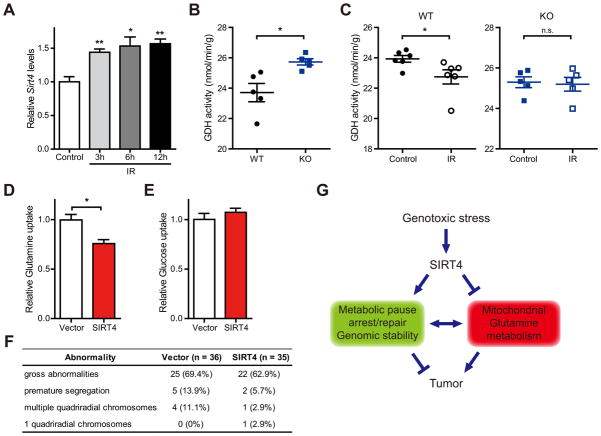
Because sirtuins regulate both cellular metabolism and stress responses (Finkel et al., 2009; Schwer and Verdin, 2008), we examined whether sirtuins were involved in the metabolic adaptation to DNA damage. We first examined the expression of sirtuins in the response to DNA damage. Specifically, we probed SIRT1, which is involved in stress responses (Haigis and Guarente, 2006), as well as mitochondrial sirtuins (SIRT3–5), which have been shown to regulate amino acid metabolism (Haigis et al., 2006; Hallows et al., 2011; Nakagawa et al., 2009). Remarkably, SIRT4 mRNA levels were induced by nearly 15-fold at 15 hr after CPT treatment and 5-fold after etoposide (ETS), a topoisomerase 2 inhibitor, in HEK293T cells (Figure 2A). Interestingly, the induction of SIRT4 was significantly higher than the induction of SIRT1 and mitochondrial SIRT3 (~2-fold), sirtuins known to be induced by DNA damage and regulate cellular responses to DNA damage (Sundaresan et al., 2008; Vaziri et al., 2001; Wang et al., 2006). Moreover, overall mitochondrial mass was increased by only 10% in comparison with control cells (Figure S2A), indicating that the induction of SIRT4 is not an indirect consequence of mitochondrial biogenesis. These data hint that SIRT4 may have an important, previously undetermined role in the DDR.
Figure 2. SIRT4 is induced by DNA damage stimuli.
(A) Relative mRNA expression levels of indicated sirtuins in HEK293T cells treated with 14 μM CPT or 25 μM ETS for 15 hr were measured by qRT-PCR (n = 4). β-ACTIN was used as an endogenous control for qRT-PCR.
(B) Relative Sirt4 mRNA levels in primary MEFs at 12 hr after treatment with CPT (14 μM), ETS (25 μM), IR (5 Gy) or UV (30 J/m2) were measured by qRT-PCR (n = 3–4). β-Actin was used as an endogenous control for qRT-PCR.
(C) SIRT4 protein in whole cell lysates from HEK293T cells treated with CPT (14 μM) or ETS (25 μM) for 15 hr was detected by immunoblotting with anti-human SIRT4. β-ACTIN serves as a loading control.
(D) SIRT4 protein in transformed WT and SIRT4 KO MEFs treated CPT (14 μM) for the indicated times. β-Actin serves as a loading control.
Data are means ±SEM. *p < 0.05, **p < 0.005, ***p < 0.0001. All experiments were performed at least three times in duplicate.
See also Figure S2.
To test the induction of SIRT4 in the general genotoxic stress response, we treated cells with other types of DNA damage, including UV and gamma-irradiation (IR). SIRT4 mRNA levels were also increased by these genotoxic agents (Figures S2B and S2C) and low doses of CPT and UV treatment also induced SIRT4 expression (Figures S2D and S2E). We observed similar results with MEFs (Figures 2B and 2D; Figure S2F) and HepG2 cells (Figure S2G). DNA damaging agents elevated SIRT4 in p53-inactive HEK293T cells (Figures 2A and 2C) and in p53-null PC3 human prostate cancer cells (Figure S2H), suggesting that SIRT4 can be induced in a p53-independent manner.
To examine whether the induction of SIRT4 occurred as a result of cell cycle arrest, we measured SIRT4 levels after the treatment of nocodazole, which inhibits microtubule polymerization to block mitosis. While treatment with nocodazole completely inhibited cell proliferation (data not shown), SIRT4 expression was not elevated (Figure S2I). In addition, we analyzed SIRT4 expression in distinct stages of the cell cycle in HepG2 cells synchronized with thymidine block (Figure S2J, Left). SIRT4 mRNA levels were measured at different times after release and were not elevated during G1 or G2/M phases (Figure S2J, Right), suggesting that SIRT4 is not induced as a general consequence of cell cycle arrest. Next, we re-examined the localization of SIRT4 after DNA damage. SIRT4 localizes to the mitochondria of human and mouse cells under basal, unstressed conditions (Ahuja et al., 2007; Haigis et al., 2006). Following CPT treatment, SIRT4 colocalized with MitoTracker, a mitochondrial-selective marker, indicating that SIRT4 retains its mitochondrial localization after exposure to DNA damage (Figure S2K). Taken together, our findings demonstrate that SIRT4 is induced by multiple forms of DNA damage in numerous cell types, perhaps to coordinate the mitochondrial response to genotoxic stress.
SIRT4 represses glutamine anaplerosis
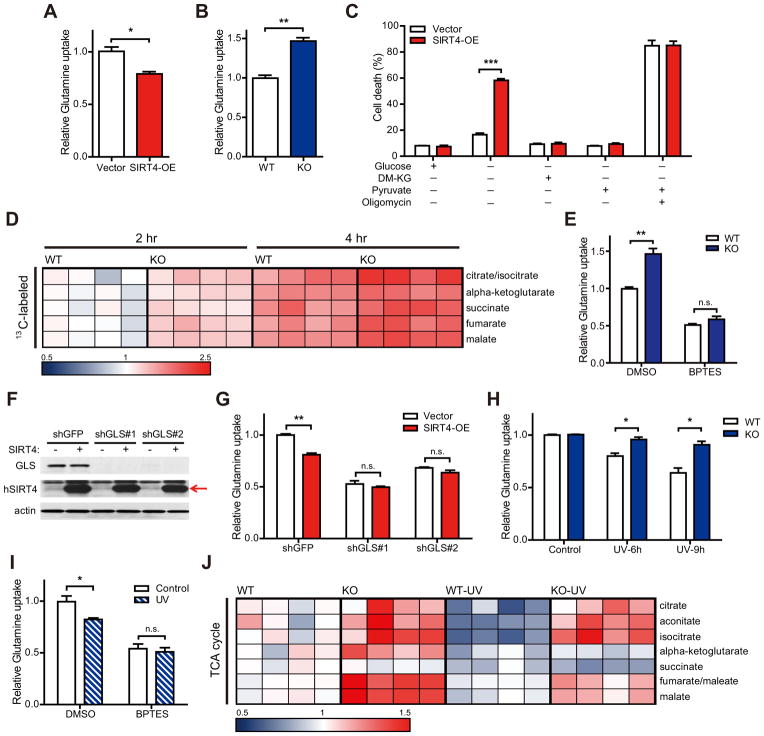
We observed that glutamine anaplerosis is repressed by genotoxic stress (Figure 1) and SIRT4 is induced by DNA damage (Figure 2). Additionally, previous studies reported that SIRT4 represses glutamine anaplerosis (Haigis et al., 2006). We next tested whether SIRT4 directly regulates cellular glutamine metabolism and contribution of glutamine to the TCA cycle. Like DNA damage, SIRT4 overexpression (SIRT4-OE) in HepG2, HeLa or HEK293T cells resulted in the repression of glutamine consumption (Figure 3A; Figures S3A–C). Conversely, SIRT4 knockout (KO) MEFs consumed more glutamine than did wild-type (WT) cells (Figure 3B).
Figure 3. SIRT4 represses mitochondrial glutamine metabolism in response to DNA damage.
(A and B) Glutamine uptake in HepG2 cells stably expressing empty vector (Vector) or SIRT4 (SIRT4-OE) (A) or in immortalized WT and SIRT4 KO MEFs (B) (n = 3).
(C) HEK293T Vector or SIRT4-OE cells deprived of glucose were given DM-KG (7 mM), pyruvate (1 mM), and/or oligomycin (5 μg/ml). Cell viability was measured via propidium iodide (PI) exclusion assay (n = 3).
(D) Relative abundance of 13C-labeled TCA cycle intermediates (M+5 for α-ketoglutarate or M+4 for others) to the unlabeled from transformed WT and SIRT4 KO MEFs at the indicated times after pulse of 13C-glutamine (n = 4 samples of each condition).
(E) Glutamine uptake in immortalized WT and SIRT4 KO MEFs treated with dimethyl sulfoxide (DMSO) or BPTES (10 μM) (n = 3–4).
(F) GLS1 protein levels in HEK293T cells expressing GLS1-specific (shGLS#1 and 2) or control (shGFP) shRNAs. β-ACTIN serves as a loading control.
(G) Glutamine uptake in control (shGFP) or GLS-knockdown (shGLS#1 and 2) cells after transfection with empty vector (Vector ) and SIRT4 (SIRT4-OE) (n = 3).
(H) Relative glutamine uptake to each control in transformed WT and SIRT4 KO MEFs measured at the indicated times after 20 J/m2 UV exposure (n = 3).
(I) Glutamine uptake in DMSO or BPTES (10 μM) treated immortalized MEFs after 20 J/m2 UV exposure (n = 3).
(J) Heat map comparing relative levels of TCA cycle intermediates in transformed WT and SIRT4 KO MEFs at 4 hr after 20 J/m2 UV exposure (n = 4).
Data are means ±SEM. n.s., not significant. *p < 0.05, **p < 0.005, ***p < 0.0001.
See also Figure S3.
Mitochondrial glutamine catabolism refuels the TCA cycle and is essential for viability in the absence of glucose (Choo et al., 2011; Yang et al., 2009). Thus, we examined the effect of SIRT4 on cell survival during glucose deprivation. Overexpression of SIRT4 in HEK293T or HeLa cells increased cell death in glucose-free media compared to control cells (Figure 3C; Figure S3D). Importantly, this cell death was completely rescued by the addition of pyruvate or cell permeable dimethyl α-ketoglutarate (DM-KG), demonstrating that SIRT4 overexpression reduced the ability of cells to utilize glutamine for mitochondrial energy production. Moreover, cell death was equally maximized in the absence of glucose and presence of the mitochondrial ATPase inhibitor oligomycin (Figure 3C). These findings are in line with the model that SIRT4 induction with DNA damage limits glutamine metabolism and utilization by the TCA cycle.
We next utilized a metabolomic approach to interrogate glutamine usage in the absence of SIRT4. SIRT4 KO MEFs demonstrated elevated levels of TCA cycle intermediates (Figure 3J, WT versus KO), whereas intermediates of glycolysis were comparable with WT cells (data not shown). Nucleotides and other metabolites downstream of glutamine metabolism were not coordinately regulated by SIRT4 loss (Figure S3E and data not shown). Next, we analyzed glutamine flux in WT and SIRT4 KO MEFs in medium containing [U-13C5]glutamine for 2 or 4 hours and measured isotopic enrichment of TCA cycle intermediates. Loss of SIRT4 promoted a higher rate of incorporation of 13C-labeled metabolites derived from [U-13C5]glutamine in all TCA cycle intermediates measured (Figure 3D). These data provide direct evidence that SIRT4 loss drives increased entry of glutamine-derived carbon into the TCA cycle.
Next, we examined the mechanisms involved in this repression of glutamine anaplerosis. GLS is the first required enzyme for mitochondrial glutamine metabolism (Curthoys and Watford, 1995) and its inhibition limits glutamine flux into the TCA cycle (Wang et al., 2010; Le et al., 2012; Yuneva et al., 2012). Treatment with bis-2-(5-phenylacetoamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) (Robinson et al., 2007), an inhibitor of GLS1, repressed glutamine uptake and completely rescued the increased glutamine consumption of SIRT4 KO cells (Figure 3E). Moreover, SIRT4 overexpression no longer inhibited glutamine uptake when GLS1 was reduced by using short hairpin RNAs (shRNAs) (Figures 3F and 3G), demonstrating that SIRT4 regulates mitochondrial glutamine metabolism. SIRT4 is a negative regulator of GDH activity (Haigis et al., 2006) and SIRT4 KO MEFs exhibited increased GDH activity in comparison with WT MEFs (Figure S3F). To test whether SIRT4 regulates mitochondrial glutamine metabolism via inhibiting GDH activity, we measured glutamine uptake in WT and SIRT4 KO cells in the presence of EGCG, a GDH inhibitor (Choo et al., 2011; Li et al., 2006). The treatment of EGCG partially rescued the increased glutamine uptake of KO cells (Figure S3G), suggesting that GDH contributes to the role of SIRT4 in glutamine metabolism.
SIRT4 represses mitochondrial glutamine metabolism after DNA damage
The induction of SIRT4 after DNA damage and regulation of glutamine metabolism by SIRT4 led us to speculate that SIRT4 may repress glutamine anaplerosis in response to DNA damage. Thus, we measured cellular glutamine consumption after UV exposure with transformed and non-transformed WT and SIRT4 KO MEFs. As expected, UV treatment suppressed glutamine uptake in WT cells (Figure 3H; Figure S3H). Strikingly, we found that KO cells were unable to repress glutamine uptake in response to DNA damage (Figure 3H; Figure S3H). We tested the involvement of glutamine anaplerosis by treated cells with chemical inhibitors of GLS1. Intriguingly, UV treatment could not further repress glutamine uptake in the presence of BPTES (Figure 3I), indicating that DNA damage inhibits mitochondrial glutamine metabolism. In addition, we observed similar results with the compound 968, a small molecule inhibitor of GLS1 (Figure S3I) (Wang et al., 2010).
To probe further whether SIRT4 limits glutamine utilization to repress the TCA cycle after DNA damage, we performed metabolomic analysis with WT and SIRT4 KO MEFs with or without UV exposure. The levels of several TCA cycle intermediates remained elevated in KO cells compared to WT cells after UV exposure (Figure 3J), corroborating the idea that SIRT4 is required for the proper repression of mitochondrial glutamine metabolism in response to DNA damage.
SIRT4 regulates cell cycle progression and genomic fidelity in response to DNA damage
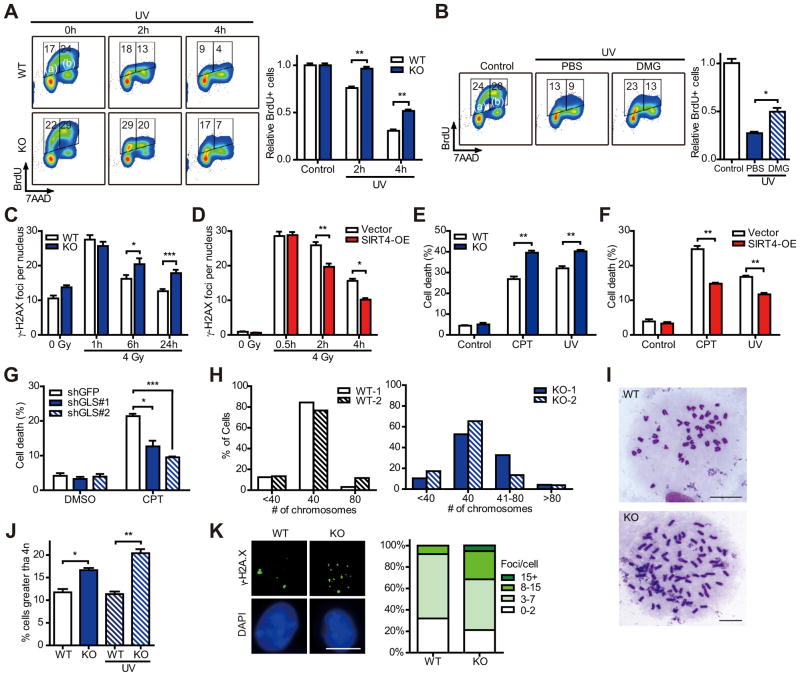
DNA damage initiates cell cycle arrest, which is crucial for maintenance of genomic integrity and ultimately for the prevention of cancer (Ciccia and Elledge, 2010). Glutamine is an essential metabolite for proliferation (Gaglio et al., 2009; Jones and Thompson, 2009; Wise and Thompson, 2010) and required for transition from G1 to S phase (Colombo et al., 2011). Because our studies demonstrate that SIRT4 is induced by DNA damage to repress the utilization of glutamine, we interrogated the role of SIRT4 in cell cycle progression after DNA damage. To induce DNA damage independently of the rate of proliferation, we treated WT and SIRT4 KO MEFs with UV radiation and then pulsed with bromodeoxyuridine (BrdU) for 30 min followed by staining with anti-BrdU FITC and 7-AAD. As expected, the population of the BrdU labeled [BrdU+ (S phase), (a) + (b)] cells was decreased in WT cells in a time-dependent manner (Figure 4A; Figure S4A). However, this inhibition of BrdU incorporation was significantly delayed in KO MEFs, and this difference was most noticeable in cells in early S phase (Figure 4A, (a)). These cells were in G1 phase at the time of UV treatment and entered S phase during pulsing with BrdU. We observed similar results with IR treatment (Figure S4B). At 6 hr after IR treatment, G1 phase cells were nearly absent in KO cells but still present in WT cells (Figure S4B, red circle).
Figure 4. SIRT4 is involved in cellular DNA damage responses.
(A) The measurement of BrdU+ cells and total DNA content in transformed WT and SIRT4 KO MEFs at the indicated times after 20 J/m2 UV exposure (n = 3).
(B) The BrdU+ cell and total DNA content in transformed WT MEFs incubated with PBS or DMG (10 mM) after UV exposure (n = 3).
(C and D) The number of γ-H2AX foci in immortalized WT and SIRT4 KO MEFs (C), or Vector and SIRT4-OE HeLa cells (D) was counted at the indicated times after IR treatment.
(E and F) Survival of immortalized WT and SIRT4 KO MEFs (E), or Vector and SIRT4-OE HepG2 cells (F) treated with or without CPT (14 μM) or UV (30 J/m2) (n = 3–4). Cell viability was measured via PI exclusion assay.
(G) Survival of HepG2 cells expressing control (shGFP) or GLS1-specific (shGLS#1 and 2) shRNAs were treated with DMSO or CPT (14 μM) for 24 hr (n = 3). Cell viability was measured via PI exclusion assay.
(H and I) The percentage of chromosome number (H) and representative images of chromosome spread (I) of WT and SIRT4 KO MEFs at passage 2. Numbers of spreads counted were 64, 61, 142 and 104 for WT-1, WT-2, KO-1 and KO-2, respectively. Scale bar represents 10 μm.
(J) Transformed WT and SIRT4 KO MEFs were treated with or without 20 J/m2 UV and the percentage of cells containing greater than 4n is analyzed by flow cytometry (n = 3).
(K) Immunofluorescent staining of transformed WT and SIRT4 KO MEFs using nuclear (DAPI) and DSBs (γ-H2AX) markers (left). Scale bar represents 10 μm. The percentage of nuclei with the indicated number of γH2AX foci (right). WT MEFs (n = 119); KO MEFs (n = 71).
Data are means ±SEM. *p < 0.05, **p < 0.005, ***p < 0.0001.
See also Figure S4.
To probe the connection between glutamine metabolism and cell cycle arrest after DNA damage, we tested whether the inhibition of DNA synthesis upon DNA damage would be affected by the addition of a downstream metabolite of glutamine. We treated cells with dimethyl-glutamate (DMG), a cell-permeable glutamate donor (Maechler and Wollheim, 1999) and measured the level of BrdU+ cells after UV treatment. To our surprise, DMG treatment promoted BrdU incorporation in response to DNA damage, as more cells entered early S phase by the addition of DMG (Figure 4B; Figure S4C, PBS versus DMG). DMG treatment did not augment normal cell cycle proliferation (Figure S4D). Although the treatment of DMG caused a mild increase in the proportion of BrdU+ cells in the KO cells, the fraction of BrdU+ cells in WT cells increased robustly upon DMG-treatment compared to PBS treatment (Figure S4E). These data suggest that reduced glutamine metabolism may limit proliferation, contributing to a metabolic checkpoint in response to genotoxic stress. Bypassing this checkpoint by adding soluble metabolites downstream of glutamine can partially promote cell cycle progression even in the presence of DNA damaging conditions.
Defects in the DDR may lead to accumulation of DNA damage and also may promote cell death. To assess whether SIRT4 affects genomic integrity in response to genotoxic stress, WT and SIRT4 KO cells were irradiated and then the number of γH2AX foci, known to localize at DSBs (Rogakou et al., 1998), was counted. The clearance of γH2AX foci was significantly impaired in KO cells (Figure 4C), while SIRT4 overexpression accelerated the clearance of foci (Figure 4D). Moreover, SIRT4 KO cells showed significantly elevated levels of cell death compared to WT cells (Figure 4E), while SIRT4 overexpression was protective (Figure 4F). We next examined survival of control and GLS1 knockdown cells against DNA damage and discovered that inhibition of mitochondrial glutamine metabolism can mirror the protective role of SIRT4 overexpression (Figure 4G), demonstrating a link between mitochondrial glutamine metabolism and the DDR.
Given the role for SIRT4 in regulating the metabolic checkpoint in response to DNA damage, we analyzed whether SIRT4 loss influenced chromosome stability. Chromosome spreads from two pairs of WT and SIRT4 KO primary MEFs revealed that KO cells possessed more aneuploidy (Figures 4H and 4I). Furthermore, more polyploidy cells were found in transformed KO cells compared to WT cells and this phenotype became more severe after UV treatment (Figure 4J). Aberrant activation of the retinoblastoma (Rb)-E2F pathway by oncogenes leads to the replication-induced DSBs and formation of γH2AX foci (Bester et al., 2011). Hence, we analyzed the formation of γH2AX foci of WT and SIRT4 KO MEFs transformed with Ras and E1A, known to inactivate Rb (Harbour and Dean, 2000), and observed elevated γH2AX foci in KO cells (Figure 4K). Taken together, findings from multiple approaches demonstrate that SIRT4 is a critical regulator of genome fidelity.
SIRT4 represses tumor proliferation
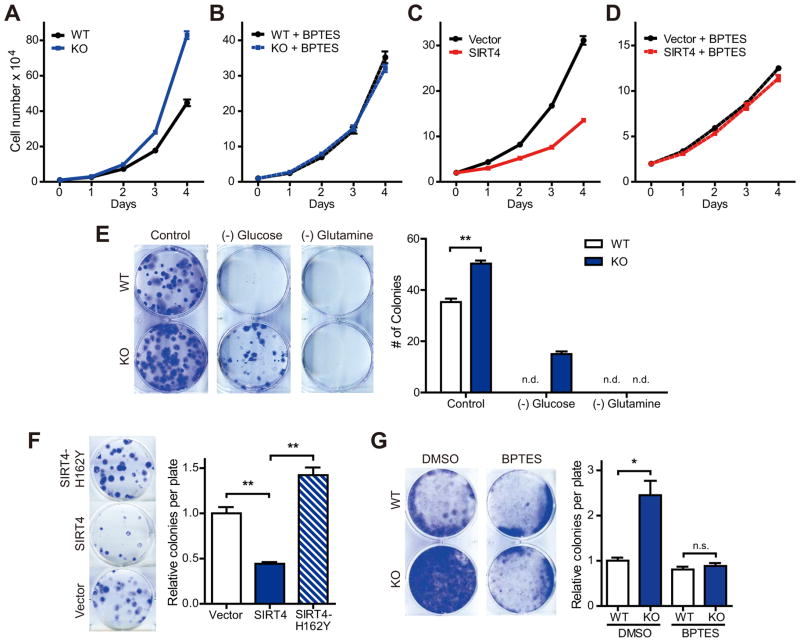
Genomic instability is one hallmark of tumorigenicity. Another feature of tumor cells is rapid cell proliferation, fueled in some cases by elevated glutamine utilization (Jones and Thompson, 2009; Wise and Thompson, 2010). Thus, we tested the idea that increased glutamine metabolism in SIRT4 KO MEFs may support proliferation. Indeed, KO cells significantly grew faster than did WT cells (Figure 5A). To test whether enhanced glutamine metabolism contributed to the proliferative phenotype of KO cells, we cultured cells with GLS1 inhibitors and measured proliferation. Remarkably, BPTES and 968 completely abrogated the increased proliferation of KO cells (Figure 5B; Figure S5A). To probe the contribution of GDH, we measured proliferation in presence of EGCG and found it likewise abrogated the proliferative phenotype of KO cells (Figure S5B). In contrast, overexpression of SIRT4 in HeLa cells, which use glutamine as a major energy source, significantly inhibited their growth (Figure 5C). Importantly, control and SIRT4-overexpressing cells proliferated at similar rates when cultured in media containing BPTES or 968 (Figure 5D; Figure S5C), highlighting the role of SIRT4 in this pathway.
Figure 5. SIRT4 has tumor suppressive function.
(A and B) Growth curves of WT and SIRT4 KO MEFs (n = 3) cultured in standard media (A) or media supplemented with BPTES (10 μM) (B). Data are means ±SD.
(C and D) Growth curves of Vector and SIRT4-OE HeLa cells (n = 3) cultured in standard media (C) or media supplemented with BPTES (10 μM) (D). Data are means ±SD.
(E) Focus formation assays with transformed WT and SIRT4 KO MEFs (left). Cells were cultured with normal medium or medium without glucose or glutamine for 10 days and stained with crystal violet. The number of colonies was counted (right) (n =3 samples of each condition). n.d., not determined.
(F) Focus formation assays with transformed KO MEFs reconstituted with SIRT4 or a catalytic mutant of SIRT4 (n = 3). Cells were cultured for 8 days and stained with crystal violet.
(G) Contact inhibited cell growth of transformed WT and SIRT4 KO MEFs cultured in the presence of DMSO or BPTES (10 μM) for 14 days (left). The number of colonies was counted (right). Data are means ±SEM. n.s., not significant. *p < 0.05, **p < 0.005. See also Figure S5.
Defects in the proper regulation of DNA damage responses can result in the accumulation of DNA lesions leading to cancer development (Lapenna and Giordano, 2009). Moreover, glutamine metabolism is critical for oncogenic transformation and cancer cell proliferation (DeBerardinis et al., 2007; Wang et al., 2010; Weinberg et al., 2010). Thus, we reasoned that SIRT4 would have the potential to suppress tumorigenesis by repressing glutamine metabolism and/or genomic instability. Thus, we assessed tumorigenic properties of transformed SIRT4 KO MEFs. KO MEFs formed more colonies than WT MEFs in colony formation assays (Figure 5E). Neither KO cells nor WT cells were able to form colonies on glutamine deficient media. In contrast, SIRT4 KO MEFs were able to form a few colonies under glucose-deprived conditions, while WT cells were not, demonstrating that SIRT4 loss facilitates glutamine utilization to support colony formation. We found that reconstitution of KO cells with SIRT4 can reverse the phenotype, whereas reconstitution with a catalytic mutant of SIRT4 cannot (Figure 5F; Figure S5D).
We next probed the contribution of glutamine anaplerosis in the transformative properties of SIRT4 KO cells. Cancer cells exhibited loss of contact inhibition of proliferation, resulting in uncontrolled cell proliferation. WT and SIRT4 KO MEFs were transformed in media supplemented with DMSO or BPTES and colony-forming activities were determined. KO cells possessed increased colony-forming activity compared to WT cells (Figure 5G). This difference was inhibited by GLS or GDH inhibitors (Figure 5G; Figure S5E), indicating that glutamine anaplerosis contributes to the tumorigenic phenotype of SIRT4 null cells.
SIRT4 represses tumor formation in vivo
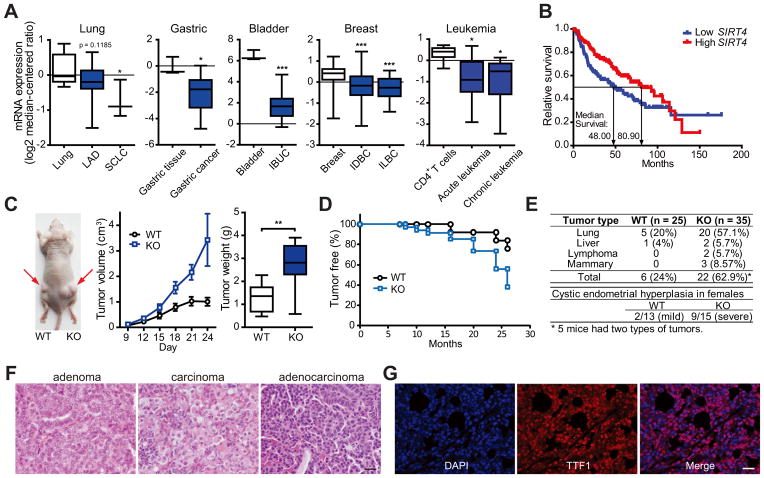
To investigate SIRT4 function in human cancers, we examined changes in SIRT4 expression. SIRT4 mRNA level was reduced in several human cancers, such as small cell lung carcinoma (Garber et al., 2001), gastric cancer (Wang et al., 2012), bladder carcinoma (Blaveri et al., 2005), breast cancer (TCGA) and leukemia (Choi et al., 2007) (Figure 6A). Of note, lower SIRT4 expression associated with shorter time to death in lung tumor patients (Shedden et al., 2008) (Figure 6B). Overall the expression data is consistent with the model that SIRT4 may play a tumor suppressive role in human cancers.
Figure 6. SIRT4 is a mitochondrial tumor suppressor.
(A) SIRT4 mRNA levels were determined using the Oncomine microarray database (http://www.oncomine.org) in normal versus lung, gastric, bladder, breast cancer and leukemia. LAD, lung adenocarcinoma; SCLC, small cell lung carcinoma; IBUC, infiltrating bladder urothelial carcinoma; IDBC, invasive ductal breast carcinoma; ILBC, invasive lobular breast carcinoma. The boxes represent the interquartile range; whiskers represent the 10th–90th percentile range; bars represent the median.
(B) Kaplan-Meier curve comparing time to survival between lung adenocarcinomas with the lowest (< 25th percentile) versus highest (> 25th percentile) SIRT4 expression was determined using the Oncomine database. p = 0.0354, log-rank test.
(C) Representative image of tumors resulting from xenograft with transformed WT and SIRT4 KO MEFs. Tumor volume and weight were measured (n = 8 tumors/genotypes). The boxes represent the interquartile range; whiskers represent the 10th–90th percentile range; bars represent the median.
(D and E) Tumor-free curve (D) and analysis of tumor types (E) in WT and SIRT4 KO mice. p = 0.0035, log-rank test.
(F) Histological sections of representative lung tumors from SIRT4 KO mice with H&E staining. Scale bar represents 20 μm.
(G) Immunofluorescent staining of a representative lung adenocarcinoma from SIRT4 KO mice using nuclear (DAPI) and lung (TTF1) markers. Scale bar represents 20 μm.
Data are means ±SEM. *p < 0.05, **p < 0.005, ***p < 0.0001.
See also Figure S6.
To extend the cellular findings to in vivo models, we used multiple approaches. First we performed allograft tumor formation assays in nude mice using transformed MEFs. Accordant with cellular models, loss of SIRT4 promoted larger tumor volume and weight compared to WT tumors in the recipient mice (Figure 6C). We next examined spontaneous tumor formation in two independent SIRT4 KO mouse strains, including in a new strain of whole-body SIRT4 KO mice, generated by deleting exons 3 and 4 (Figure S6A). Like the previously reported SIRT4 KO model (Haigis et al., 2006), these SIRT4 KO mice demonstrated normal development and size. Strikingly, SIRT4 loss increased spontaneous tumor incidence throughout life (Figure 6D). Approximately 63% (22/35) of the Sirt4 null animals developed several types of tumors, most frequently lung tumors at 18–26 months of age, while 20% (5/25) of aged WT littermates developed lung tumors (Figure 6E; Figure S6B). Lung tumors from SIRT4 KO mice were categorized according to tumor types, revealing 41% (7/17) adenomas, 41% (7/17) carcinomas and 18% (3/17) adenocarcinomas (Figure 6F). To further characterize these tumors, we performed immunohistochemistry to detect thyroid transcription factor-1 (TTF1), a transcription factor expressed specifically in epithelial cells of the thyroid and lung, providing a clinical marker in the diagnosis of tumors of lung origin (Kendall et al., 2007; Weir et al., 2007). Lung tumors from SIRT4 KO mice were positive for TTF1 (Figure 6G). Moreover, we found that 60% (9/15) of female Sirt4 null animals also developed cystic endometrial hyperplasia while only 15% (2/13) WT mice exhibited mild endometrial hyperplasia (Figure 6E).
The previously reported strain of SIRT4 KO mice (Haigis et al., 2006), demonstrated the same phenotype; these SIRT4 KO animals developed lung tumors (45.5%) more frequently than WT mice (8.3%) between 18 and 22 months of age (Figure S6C). Thus, two independently derived strains of SIRT4 KO mice possessed increased spontaneous lung tumor incidence.
SIRT4 regulates glutamine metabolism in lung tissue
To test further the biological relevance of this pathway in lung, we examined whether SIRT4 is induced in vivo after exposure to DNA damaging IR treatment. Remarkably, Sirt4 was significantly induced in lung tissue after IR exposure (Figure 7A). We next examined whether IR repressed glutamine metabolism in vivo, as observed in cell culture by examining GDH activity in lung tissue from WT and SIRT4 KO mice with or without IR exposure. GDH activity was elevated in lung tissue extracts from SIRT4 KO mice compared with WT lung tissue (Figure 7B). Importantly, GDH activity was significantly decreased in lung tissue from WT mice after IR exposure, whereas not in lung tissue from KO mice (Figure 7C). Thus, these findings recapitulate our cellular studies and are in line with the model that SIRT4 induction with DNA damage limits mitochondrial glutamine metabolism and utilization.
Figure 7. SIRT4 inhibits mitochondria glutamine metabolism in vivo.
(A) Relative Sirt4 mRNA levels in lung tissues for the indicated times after whole-body IR (10 Gy) were determined by qRT-PCR (n = 3). β-Actin was used as an endogenouse control for qRT-PCR.
(B) GDH activity in lung tissue extracts from WT and SIRT4 KO mice (n = 5 animals/genotype).
(C) GDH activity in lung tissue extracts from WT (left) and SIRT4 KO (right) mice at 12 hr after whole-body IR (10 Gy) (n = 5–6 mice of each condtion).
(D and E) Glutamine (D) and glucose (E) uptake in SIRT4 KO lung tumor cells reconstituted with SIRT4 (n = 3).
(F) Chromosomal abnormalities were examined in SIRT4 KO lung tumor cells reconstituted with SIRT4 after IR (5 Gy) treatment.
(G) A proposed model illustrating the regulation of metabolic response to DNA damage by SIRT4.
Data are means ±SEM. n.s., not significant. *p < 0.05, **p < 0.005. See also Figure S7.
To assess whether the functions of SIRT4 can be reproduced in these lung tumors, cells derived from SIRT4 KO lung tumors were reconstituted with wild type SIRT4 (Figure S7A). As expected, SIRT4 reconstitution reduced glutamine uptake, but not glucose uptake (Figures 7D and 7E) and repressed proliferation (Figure S7B) of lung tumor cells. Finally, we tested whether SIRT4 repressed genomic instability in these tumor cells after DNA damage. When we examined the chromosome abnormalities after irradiation, the reconstituted tumor cells with SIRT4 exhibit decreased genomic instability (Figure 7F). Taken together, these results provide critical evidence that SIRT4 regulates both glutamine metabolism and genomic instability in tumor cells, and that loss of this critical regulatory node contributes to cancer susceptibility.
DISCUSSION
Here, we report that SIRT4 has an important role in cellular metabolic response to DNA damage by regulating mitochondrial glutamine metabolism with important implication for the DDR and tumorigenesis. First, we discovered that DNA damage represses cellular glutamine metabolism (Figure 1). Next, we found that SIRT4 is induced by genotoxic stress (Figure 2) and is required for the repression of mitochondrial glutamine metabolism (Figure 3). This metabolic response contributes to the control of cell cycle progression and the maintenance of genomic integrity in response to DNA damage (Figure 4). Loss of SIRT4 increased glutamine-dependent tumor cell proliferation and tumorigenesis (Figure 5). In mice, SIRT4 loss resulted in spontaneous tumor development (Figure 6). We demonstrate that SIRT4 is induced in normal lung tissue in response to DNA damage where it represses GDH activity. Finally, the glutamine metabolism-genomic fidelity axis is recapitulated in lung tumor cells derived from SIRT4 KO mice via SIRT4 reconstitution (Figure 7). Our studies therefore uncover SIRT4 as a important regulator of cellular metabolic response to DNA damage that coordinates repression of glutamine metabolism, genomic stability and tumor suppression.
The DDR is a highly orchestrated and well-studied signaling response that detects and repairs DNA damage. Upon sensing DNA damage, the ATM/ATR protein kinases are activated to phosphorylate target proteins, leading to cell cycle arrest, DNA repair, transcriptional regulation and initiation of apoptosis (Ciccia and Elledge, 2010; Su, 2006). Dysregulation of this pathway is frequently observed in many tumors. Emerging evidence has suggested that cell metabolism also plays key roles downstream of the DDR-induced pathways. For example, ATM has been reported to repress the rapamycin-sensitive mammalian target of rapamycin (mTORC1) pathway by activating the serine/threonine kinase LKB1/AMP-activated protein kinase (AMPK) metabolic pathway (Alexander et al., 2010). Several studies also indicate that AMPK is activated downstream of p53 via p53 target genes; sestrin 1 and sestrin 2 (Shackelford and Shaw, 2009). Moreover, damaged cells upregulate the PPP to facilitate DNA repair by generating precursors for nucleotide biosynthesis (Cosentino et al., 2011). The role of SIRT4 in the metabolic response to genotoxic stress synergizes with changes in cellular signaling pathways. SIRT4 contributes to this stress response by repressing glutamine entry into TCA cycle.
How does diminished glutamine metabolism regulate cellular responses to genotoxic stress? Proliferating cells use precursors derived from TCA cycle intermediates to synthesize NADPH, lipids, proteins and nucleic acids (DeBerardinis et al., 2008; Wise et al., 2010). For example, mitochondrial citrate is exported to the cytosol and is used for lipid synthesis and protein acetylation. However, continuous export of TCA cycle intermediates would result in the loss of mitochondrial integrity by stalling the TCA cycle, limiting anabolic pathways, and decreasing substrates for cellular respiration. Thus, refilling the mitochondrial carbon pool by glutamine is essential for the maintenance of mitochondrial integrity and its biosynthetic roles, as has been previously demonstrated (DeBerardinis et al., 2007). We find that this pathway is repressed in response to cellular stress, and additionally demonstrate that SIRT4 regulates glutamine anaplerosis.
Although our current work highlights the regulation of glutamine anaplerosis by SIRT4, other glutamine metabolism pathways may also be important for the DDR and tumorigenesis. For example, glutamine contributes to de novo synthesis of glutathione (GSH), an intracellular antioxidant that plays a critical role in cellular defense against oxidative stress (Lu, 2009). Our studies did not find a significant difference in the levels of GSH and GSH dimers (GSSG) and no measurable flux of glutamine into GSH under these conditions and at short time points after damage (data not shown). Glutamine metabolism also contributes to lipid synthesis, nucleotide synthesis and NADPH levels. Sirtuins have several targets and synergically regulate the same pathway. For example, SIRT3 has been shown to bind and regulate multiple enyzmes in mitochondrial fatty-acid oxidation (Hallows et al., 2011; Hirschey et al., 2010). Thus, it will be interesting for future studies to examine systematically whether SIRT4 regulates mitochondrial glutamine metabolism and other branches of glutamine metabolism via other targets.
This study identifies a important role for SIRT4 in suppressing tumor growth using a combination of human data, and cellular and mouse experiments. In humans, SIRT4 mutations have been identified colon, lung, and uterine carcinomas (http://www.cbioportal.org). Moreover, SIRT4 expression is decreased in human tumors and correlates with prognosis in lung cancer patients. Our cellular and animal studies also reveal that SIRT4 loss increases tumorigenesis, importantly in a glutamine-dependent manner. These findings also share parallels with the role of SIRT3, another mitochondrial sirtuin, in tumorigenesis. Like SIRT3, the tumors in SIRT4 KO mice arise in an age-dependent manner - after 1 year. Thus, as with SIRT3, SIRT4 loss on its own may not be the initiating event in tumorigenesis, but its loss appears to create a tumor permissive environment to promote both increased genomic instability and anabolic growth, supporting tumor cell survival and proliferation.
Several other sirtuins have been shown to play critical roles in genome maintenance and tumorigenesis. For example, SIRT1- and SIRT6-deficient cells showed an impaired DSB repair and SIRT2, SIRT3 and SIRT6 KO mice exhibit spontaneous genomic instability (Kim et al., 2010; Mao et al., 2011; Mostoslavsky et al., 2006; Oberdoerffer et al., 2008; Wang and Tong, 2009). As sirtuins are important modulators of cell metabolism (Haigis and Guarente, 2006; Houtkooper et al., 2012), it will be interesting to examine whether these sirtuins coordinately regulate genomic fidelity and tumorigenesis, in part via modulating fuel switching.
In sum, our studies reveal an important role for the mitochondrial sirtuin, SIRT4 in cancer biology and connect two hallmarks of tumor cells: genomic instability and dysregulation of glutamine metabolism. Given the importance of metabolism in cellular proliferation and tumorigenesis, these finding hold profound implications for understanding the normal metabolic response to stress, as well as for the development of cancer therapy.
EXPERIMENTAL PROCEDURES
Animal studies
Animal studies were performed according to protocols approved by the Institutional Animal Care and Use Committee, the Standing Committee on Animals at Harvard. Age-matched SIRT4 WT and KO mice were sacrificed and subjected to pathological examination. For xenograft studies, 106 transformed SIRT4 WT or KO MEFs in matrigel (BD Bioscience) for a total volume of 100 μl were injected subcutaneously in right and left flanks of 8-week-old male nude mice (Charles River). Visible tumor volume was measured on the indicated days with calipers. At the termination of the experiment, tumors were excised and weighed. For generation of SIRT4 mutant mice, chimeric mice were mated with NIH Black Swiss females (Taconic) to screen for germline transmission. Male mice bearing germline transmission were mated with female FVB EIIa-Cre mice to generate whole body exons 3 and 4 deletion of the Sirt4 gene. All experiments were approved by the Animal Care and Use Committee of the National Institute of Diabetes, Digestive and Kidney Diseases (ACUC, NIDDK).
Histological analysis
Tumors were dissected from mice, and fixed in 10% neutral-buffered formalin (Sigma) at RT overnight, and placed in 70% ethanol for at least one day. Then the tumor tissues were dehydrated through a graded alcohol series, xylene and paraffin, and then embedded in paraffin. Sections of 5 μm were cut and stained by hematoxylin and eosin (H&E).
Flow cytometric measurement of cell death and BrdU incorporation
Cells at less than 80% confluency were treated with DNA damage agents. After treatments, cells were harvested by trypsinization, pelleted by centrifugation and resuspended in PBS containing 3% FBS. The measurement of cell death was performed by Flow cytometry using propidium iodide (PI) staining as previously described. The incorporation of BrdU into the genomic DNA was measured with BrdU Flow Kit (BD Pharmingen) according to the manufacturer’s instructions.
γH2AX immunofluorescence
Cells grown on coverslips or 8-well microscopy slides were fixed for 20 min with 4% paraformaldehyde and permeabilized by 0.1% Triton X-100 for 3min. Cells were then incubated overnight at 4 degrees with mouse monoclonal antibodies against γH2AX (Ser139) (Millipore, 1:200 dilution) followed by goat anti-mouse IgG-FITC (Santa Cruz, 1:300). Cell images were taken under a Zeiss microscope using a 63× objective and analyzed for foci/nucleus.
Glutamine and glucose Measurements
Glutamine, ammonia, glucose and lactate levels in culture media were measured using the BioProfile FLEX analyzer (Nova Biomedical) as previously described (Finley et al., 2011). Briefly, fresh media were added to a 6-well plate of cells and metabolite levels in the media were measured 6–9 hr later and normalized to the number of cells in each well.
Metabolites profiling
Metabolites were extracted and analyzed as previously described (Finley et al., 2011). Briefly, SIRT4 WT and KO MEFs were plated, then metabolites were extracted at 4hr after 20 J/m2 UV exposure. Metabolite levels were normalized to the total of all metabolites detected.
Statistical analysis
Unpaired two-tailed Student’s t tests were performed unless otherwise noted. All experiments were performed at least two or three times. For the tumor incidence and survival study, the Log-rank (Mantel-Cox) was performed.
Supplementary Material
SIGNIFICANCE.
Genomic instability and altered metabolism are key features of many cancer cells. Thus, defining the factors that are involved in regulating DNA damage responses and metabolic processes may have profound implications for the development of strategies to prevent or treat cancer. We find that SIRT4, a mitochondria-localized sirtuin, functions as a tumor suppressor by regulating metabolic responses to DNA damage and repressing mitochondrial glutamine metabolism. Importantly, we also show that SIRT4 knockout mice spontaneously develop several types of tumors and SIRT4 expression is decreased in many human cancers. Our findings suggest that SIRT4 may be a potential therapeutic target against tumors.
HIGHLIGHTS.
DNA damage induces SIRT4 and inhibits mitochondrial glutamine metabolism.
SIRT4 is required for proper repression of glutamine metabolism after DNA damage. 3. SIRT4 loss promotes genomic instability.
SIRT4 represses tumor growth and SIRT4 knockout mice develop tumors with age.
Acknowledgments
We thank Roderick T. Bronson for analyzing tumors, Moon Hee Yang for help with allograft assays, Annie Lee for technical assistance, and the Nikon Imaging Center at Harvard Medical School. We thank Kevin Haigis for critical reading of the manuscript. S.M.J. was supported in part by National Research Foundation of Korea Grant funded by the Korean Government (NRF-2010–357–C00087). M.H. is supported in part by NIH grant AG032375, the Glenn Foundation for Medical Research and American Cancer Society New Scholar Award. A.C.K is supported by the National Cancer Institute Grant R01 CA157490, Kimmel Scholar Award, and AACR-PanCAN Career Development Award. This work was also supported in part by the Intramural Research Program of the National Institute of Diabetes, Digestive and Kidney Diseases, National Institutes of Health, USA.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and seven figures
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, Maechler P, Verdin E. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282:33583–33592. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaveri E, Simko JP, Korkola JE, Brewer JL, Baehner F, Mehta K, Devries S, Koppie T, Pejavar S, Carroll P, et al. Bladder cancer outcome and subtype classification by gene expression. Clin Cancer Res. 2005;11:4044–4055. doi: 10.1158/1078-0432.CCR-04-2409. [DOI] [PubMed] [Google Scholar]
- Choi YL, Tsukasaki K, O’Neill MC, Yamada Y, Onimaru Y, Matsumoto K, Ohashi J, Yamashita Y, Tsutsumi S, Kaneda R, et al. A genomic analysis of adult T-cell leukemia. Oncogene. 2007;26:1245–1255. doi: 10.1038/sj.onc.1209898. [DOI] [PubMed] [Google Scholar]
- Choo AY, Kim SG, Vander Heiden MG, Mahoney SJ, Vu H, Yoon SO, Cantley LC, Blenis J. Glucose addiction of TSC null cells is caused by failed mTORC1-dependent balancing of metabolic demand with supply. Mol Cell. 2011;38:487–499. doi: 10.1016/j.molcel.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo SL, Palacios-Callender M, Frakich N, Carcamo S, Kovacs I, Tudzarova S, Moncada S. Molecular basis for the differential use of glucose and glutamine in cell proliferation as revealed by synchronized HeLa cells. Proc Natl Acad Sci U S A. 2011;108:21069–21074. doi: 10.1073/pnas.1117500108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosentino C, Grieco D, Costanzo V. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J. 2011;30:546–555. doi: 10.1038/emboj.2010.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curthoys NP, Watford M. Regulation of glutaminase activity and glutamine metabolism. Annu Rev Nutr. 1995;15:133–159. doi: 10.1146/annurev.nu.15.070195.001025. [DOI] [PubMed] [Google Scholar]
- Dang CV. Glutaminolysis: supplying carbon or nitrogen or both for cancer cells? Cell Cycle. 2010;9:3884–3886. doi: 10.4161/cc.9.19.13302. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng CX. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res. 2006;34:1416–1426. doi: 10.1093/nar/gkl010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio D, Soldati C, Vanoni M, Alberghina L, Chiaradonna F. Glutamine deprivation induces abortive s-phase rescued by deoxyribonucleotides in k-ras transformed fibroblasts. PLoS One. 2009;4:e4715. doi: 10.1371/journal.pone.0004715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Hallows WC, Yu W, Smith BC, Devries MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, Gerald WL, Powers S, Mu D. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci U S A. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov. 2009;8:547–566. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- Li C, Allen A, Kwagh J, Doliba NM, Qin W, Najafi H, Collins HW, Matschinsky FM, Stanley CA, Smith TJ. Green tea polyphenols modulate insulin secretion by inhibiting glutamate dehydrogenase. J Biol Chem. 2006;281:10214–10221. doi: 10.1074/jbc.M512792200. [DOI] [PubMed] [Google Scholar]
- Maechler P, Wollheim CB. Mitochondrial glutamate acts as a messenger in glucose-induced insulin exocytosis. Nature. 1999;402:685–689. doi: 10.1038/45280. [DOI] [PubMed] [Google Scholar]
- Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MM, McBryant SJ, Tsukamoto T, Rojas C, Ferraris DV, Hamilton SK, Hansen JC, Curthoys NP. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) Biochem J. 2007;406:407–414. doi: 10.1042/BJ20070039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TT. Cellular responses to DNA damage: one signal, multiple choices. Annu Rev Genet. 2006;40:187–208. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol Cell Biol. 2008;28:6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, Nemoto S, Finkel T, Gu W, Cress WD, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8:1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- Wang F, Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1’s repressive interaction with PPARgamma. Mol Biol Cell. 2009;20:801–808. doi: 10.1091/mbc.E08-06-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wen YG, Li DP, Xia J, Zhou CZ, Yan DW, Tang HM, Peng ZH. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29:77–83. doi: 10.1007/s12032-010-9766-y. [DOI] [PubMed] [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Sudderth J, Dang T, Bachoo RM, McDonald JG, DeBerardinis RJ. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 2009;69:7986–7993. doi: 10.1158/0008-5472.CAN-09-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.