Fig. 2.
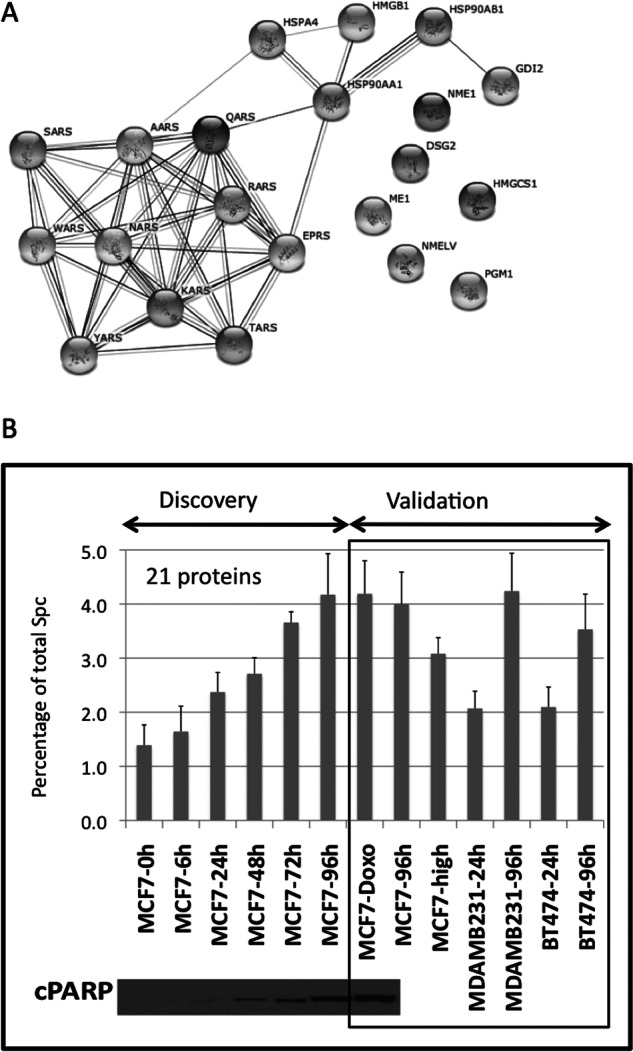
A group of secreted proteins correlates with apoptosis and can be used as a cell viability indicator. (A) Pathway analysis of the 21 selected proteins that correlate with apoptosis in the serum-deprivation kinetics experiment. The dataset was composed of two biological replicates with three technical replicates for each of the time points. The 21 selected proteins were uploaded into STRING software, mapped to gene objects and used to generate a biological network. (B) Graph representing the percentage of spectral counts coming from the stress signature proteins compared with total spectral counts of each sample during the secretome kinetics experiment in MCF7 cells. In the discovery phase the percentage of signal of the stress signature correlates with the levels of cPARP detected by Western blot. At 24h less than 1% of spectral counts comes from the stress proteins, while at 96h that signal represents more than 4%. The same subset of proteins was used to validate their correlation with cellular stress in: MCF7 cells treated with doxorubicin, an apoptotic inducer; in three different cell lines maintained in serum-free media for either 24h or 96h, and in a MCF7 high passage in which the apoptosis levels are increased versus the parental cells. The standard deviation was calculated using the aggregated MS signal of the 21 proteins, dividing it by the total MS signal in the sample LC-MS run, and then averaging the variances for the six technical replicas coming from two biological replicas.
