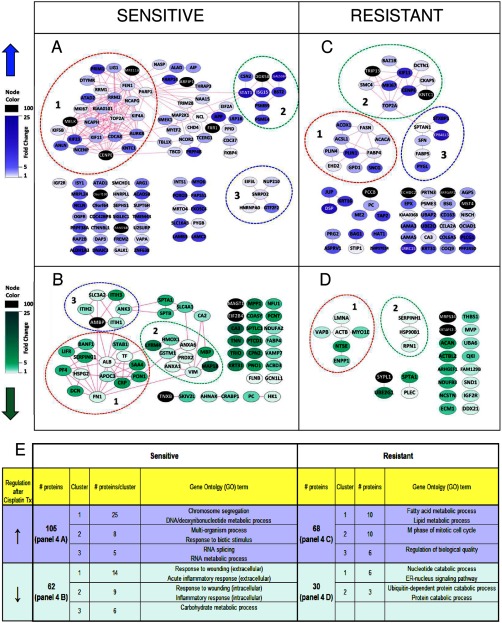
Fig. 4.
Protein-protein networks of the regulated proteins selected using a paired statistical analysis between treated and untreated conditions. The networks were generated using default settings in String (and visualized using Cytoscape. Dashed lines indicate the top three most significant clusters identified by Cluster ONE analysis. Nodes represent proteins, and the edges represent interactions that include direct (physical) and indirect (functional) associations. See Szklarczyk et al. (19) for more details on edge generation. A, up-regulated proteins in the cisplatin-sensitive tumors. B, down-regulated proteins in the cisplatin-sensitive tumors. C, up-regulated proteins in the cisplatin-resistant tumors. D, down-regulated proteins in the cisplatin-resistant tumors. E, representative GO terms identified by BiNGO analysis for the top three clusters within the regulated proteins.