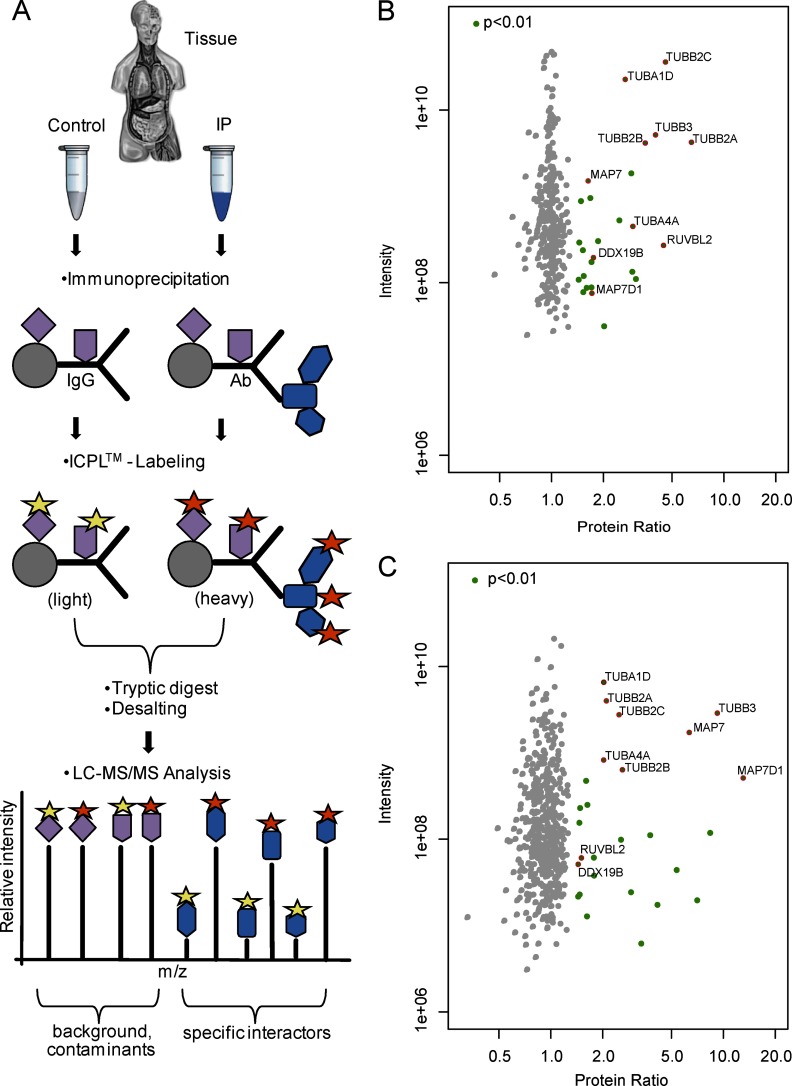
Fig. 1.
ICPL-IP: A novel approach for the quantitative protein complex analysis from native tissue. A, Experimental scheme of the ICPL-IP by using a combination of immunoprecipitation, isotope coded protein labeling, and mass spectrometry. Equal amounts of lyzed tissue are split into a control and IP. The IP contains the specific antibody, whereas the control only contains species-specific IgGs. Following IP, the samples are differently labeled (yellow and orange stars), mixed and analyzed by LC-MS/MS. After software-based analysis, nonspecific binders (purple) can be easily determined by ratios of 1:1. Specific binders to the protein of interest (blue) are detected by significant enrichment in the IP. B–C, Detection of β-tubulin protein complex components by ICPL-IP in retinal tissue. Proteins were immunoprecipitated using a monoclonal (B) or a polyclonal (C) β-tubulin antibody, respectively. Plotted are log10 ratios (x-axis) and log10 intensities (y-axis) for each quantified protein. Significantly enriched proteins in one of the two ICPL-IPs (green, p < 0.01), nonspecific binders (gray), and proteins significantly enriched in both cases (red, p < 0.01) are indicated (for details see Experimental Procedures and Table I).