Fig. 2.
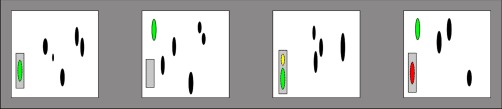
Global precision and recall computed for one feature aligned between four LC-MS maps. The ovals represent detected features. The colors represent a priori knowledge of the underlying correspondences. The green features are known to represent the same peptide based on peptide identity information. The gray boxes surrounding the features represent a feature cluster after matching; the features contained in the boxes are aligned. The size of the box represents the m/z and retention time tolerance used after retention time correction. An empty box indicates a missing value, i.e., there is no feature matched in the cluster for that file. The yellow feature is a false positive for both identity- and occurrence-based metrics, violating the assumption of a one-to-one correspondence between features. The red feature on the other hand, would be considered a false negative for the identity-based recall, as false positive for the identity-based precision, but a true positive for the occurrence-based metrics. This leads to an identity-based recall of 2/(2 + 2) = 0.5 and precision of 2/(2 + 2) = 0.5 whereas corresponding occurrence-based metrics equal 3/(3 + 1) = 0.75 and 3/(3 + 1) = 0.75, respectively. Every feature with an associated identity that is shared between all files before alignment is evaluated after the process as described above, leading to overall estimates of the alignment performance.
