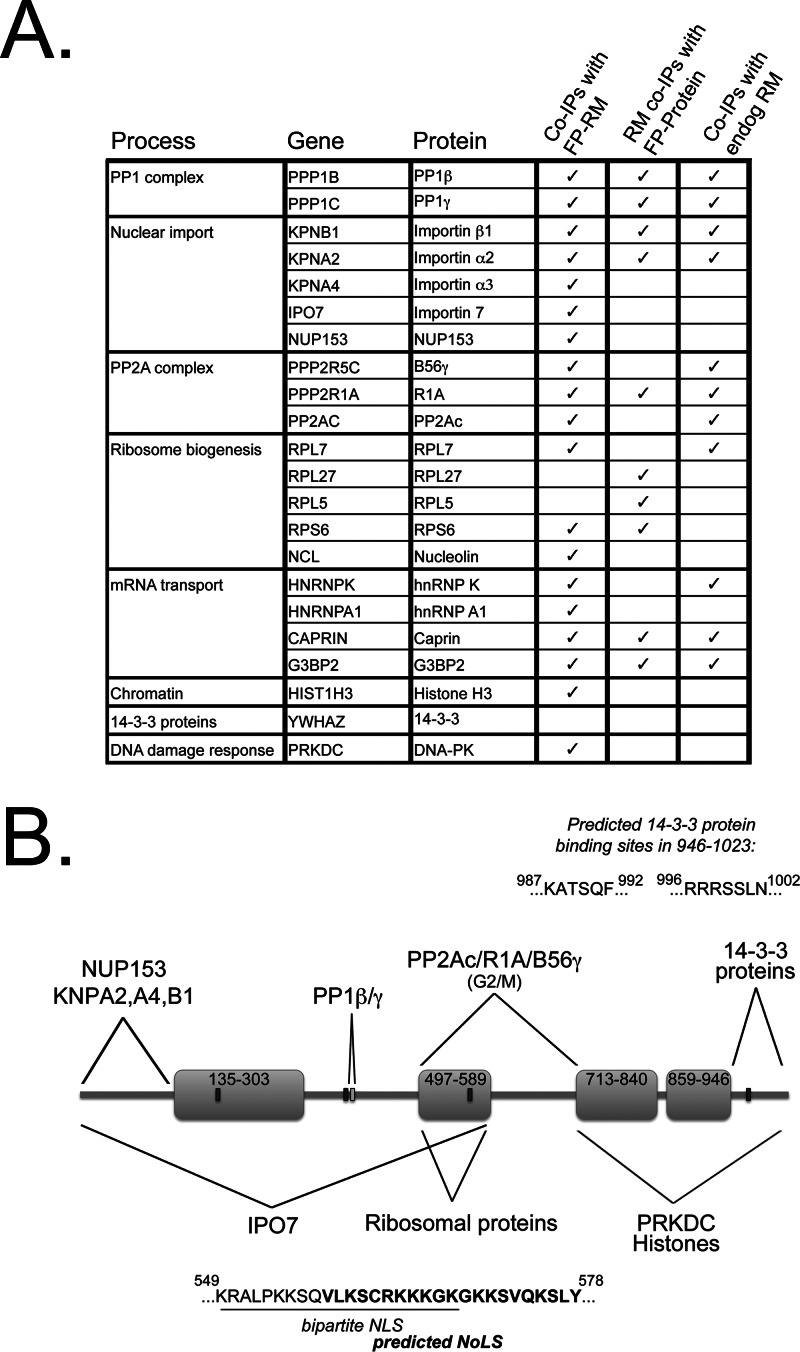
Fig. 8.
Validation summary and model of the modular interaction events mediated by specific domains of RepoMan. A, Table summarizing the validation of the RepoMan interactors discussed in this study. Where indicated, endogenous interactors were detected in pulldowns of full-length FP-tagged RepoMan (Co-IPs with FP-RM), endogenous RepoMan was detected in pulldowns of FP-tagged interactors (RM co-IPs with FP-Protein) and/or endogenous interactors were detected in pulldowns of endogenous RepoMan (Co-IPs with endog RM). With the exception of 14-3-3ζ, all of the interactors shown here validated with full-length RepoMan. B, Diagram summarizing the domain-specificity of the interactions uncovered using the “fragmentome” mapping approach. Although the extreme N terminus mediates most interactions with importins and NUPs, IPO7 (and other nonclassic importins) binds weakly to both the 1–135 and 496–591 regions of RepoMan, and more efficiently to the 1–591 fragment encompassing both of these regions. The PP1-binding RVxF domain in the N terminus mediates interaction with PP1β and PP1γ. Region 496–591, which mediates interaction with ribosomal proteins and shows additional accumulation in nucleoli, contains both a predicted bipartite nuclear localization signal (NLS, underlined) and a predicted nucleolar localization signal (NoLS, bold text). The longer 496–713 domain is required for association with PP2Ac/R1A/B56γ, and this association occurs predominantly in early mitosis. The chromatin localization pattern observed for RepoMan in interphase cells is maintained by the extreme C terminus (713–1023), which also mediates association with histone H3 and with the DNA damage response-related PRKDC kinase and 14-3-3 proteins. Deletion of the extreme C terminus (947–1023), which contains 2 predicted 14-3-3 binding sites, results in loss of 14-3-3 binding.