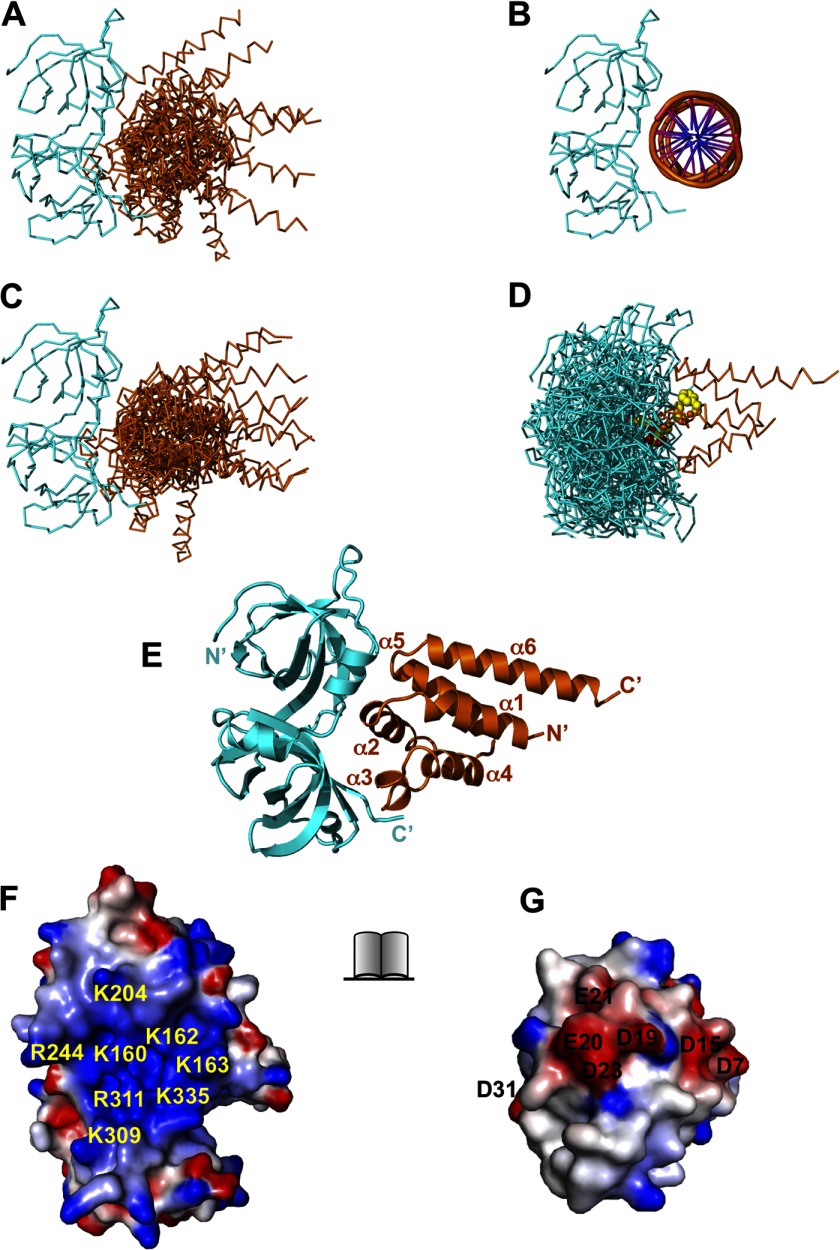
FIGURE 6.
Docking of the AIM2 PYD and HIN domains. A, superposition of the top 10 docking models of the AIM2 PYD-HIN domain complex with the HIN domain as the stationary receptor by the ClusPro server. The AIM2 PYD and HIN domain are colored orange and cyan, respectively. B, the structure of the AIM2 HIN-DNA complex (Protein Data Bank code 3RN2) is shown for comparison. C, superposition of the top 10 docking models of the AIM2 PYD-HIN complex with the PYD as the stationary receptor by the ClusPro server. The 10 models were superimposed on their HIN domains. D, the original 10 docking models for C are superimposed on their PYDs with the AIM2 residues Asp-19, Glu-20, Asp-23, Phe-27, and Phe-28 shown as yellow spheres. E, the top scoring model from A was energy-minimized with Rosetta and is shown in ribbons. The PYD and HIN domain are colored orange and cyan, respectively. The PYD helices and termini are labeled. The open book view of the PYD-HIN interface is shown as electrostatic surface in F (HIN) and G (PYD). The basic residues in HIN are labeled in yellow, and acidic residues in PYD are labeled in black.