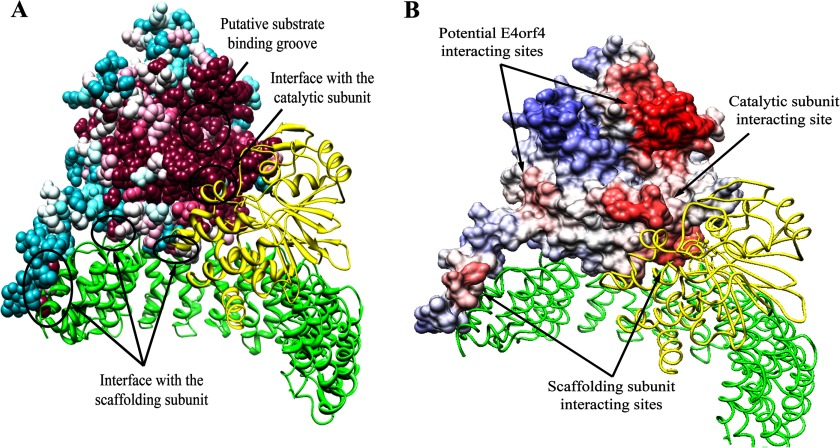
FIGURE 1.
Prediction of the B55α protein-protein interaction sites based on structure conservation. A, results of the analysis of the PP2A holoenzyme by the ConSurf program are shown. The scaffolding A and the catalytic C subunits are represented by ribbons colored green and yellow, respectively. B55α is represented by a space-filling model, colored by the following conservation scale: dark purple residues are the most conserved; white residues are the average on the conservation scale; cyan residues are variable. Known protein-protein interaction sites are marked by arrows. B, the surface of B55α was colored according to the ProMate prediction of protein-protein interaction sites as follows; residues with a higher probability for involvement in protein-protein interactions are marked in darker red, and residues with a low probability are in blue. In addition to the correctly predicted interfaces with the scaffolding (green) and catalytic (yellow) subunits, two more sites have been predicted to be involved in protein-protein interactions and are marked by arrows as potential E4orf4 interacting sites.