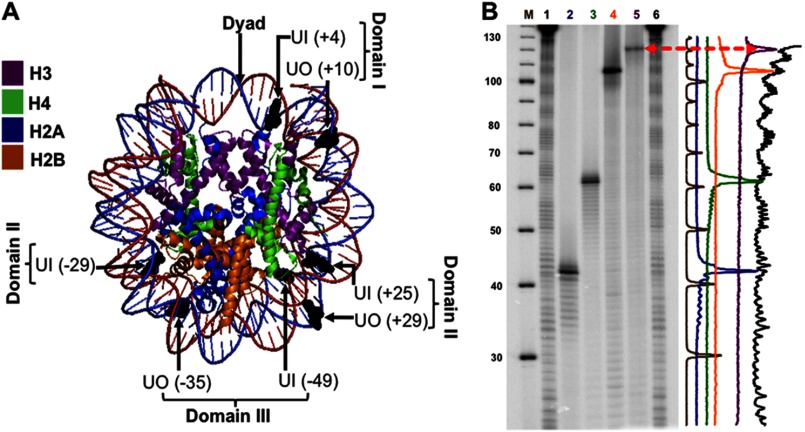
FIGURE 1.
Uracil rotational orientation and translational position in NCP substrates. A, using MacPyMOL2, the crystal structure of the 601 NCP Protein Data Bank code 3LZ0 (42) was modified to highlight the positioning of uracil (shown in black). The uracils whose DNA backbone faces the histone octamer have an in rotational orientation, and those with their DNA backbone facing the solvent are outwardly oriented (out). The translational position of the uracils was designated relative to the dyad, translational position 0, where the number in parentheses indicates the number of nucleotides toward the 5′ end (+) or toward the 3′ end (−). The flexibility of DNA increases with distance from the dyad, and thus, in terms of flexibility, domain I < II < III. B, DNA sequencing gel of hydroxyl radical footprints of the equivalent uracil-containing strand associated with the histone octamer. Lane M corresponds to the radiolabeled ([γ-32P]ATP) 10-bp DNA ladder. Lanes 1 and 6, undamaged 601-NCP treated with hydroxyl radicals. In order, lanes 2–5 show uracil-containing DNA: UO (+29), UO (+10), UO (−35), UI (−49), all treated with UDG and APE1, which cleaves 5′ to the AP site. Therefore, the location of the uracils with respect to the NCP footprint will be one nucleotide longer or shifted to the left in the intensity scan shown on the right.