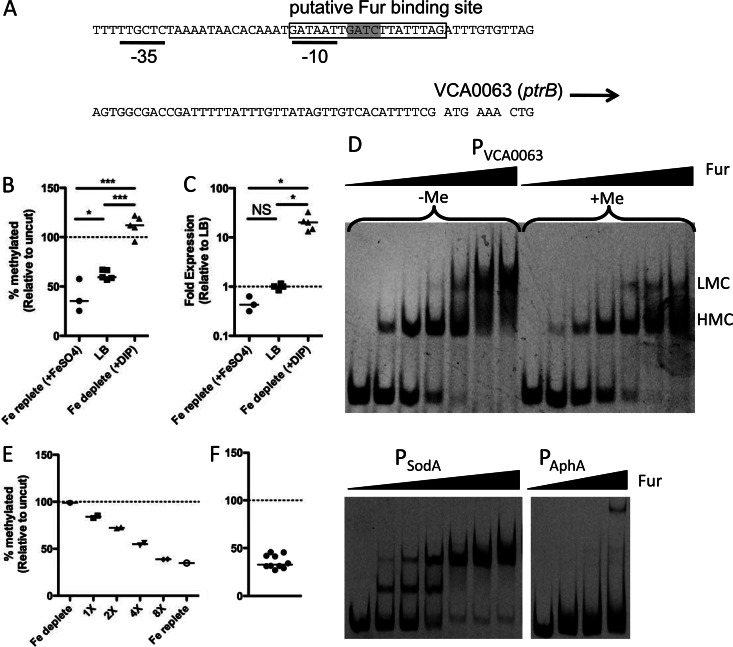
Fig 5.
Overlapping Fur and Dam methylation do not constitute an epigenetic switch at the VCA0063 promoter. (A) Genetic architecture of the VCA0063 promoter, showing the overlapping putative Fur binding site (boxed), −35 and −10 consensus sequences (black lines), and Dam site (highlighted in gray). (B and C) Cells were grown in iron-replete medium (LB plus 200 μM FeSO4), LB, or iron-depleted medium (LB plus 90 μM dipyridyl [DIP]), and the methylation level of PVCA0063 was determined by MeSR digestion and qPCR (B), while the expression level of VCA0063 was assessed by qRT-PCR (C). Each data point is from an independent biological replicate, and a horizontal line represents the median of each sample. Statistical significance was determined by two-tailed Student's t test (B) and Mann-Whitney (C). (D) EMSAs using probes of the indicated promoters and purified Fur protein. The final concentration of probe used in all EMSA reactions was 2 nM. The concentrations of Fur added in EMSAs with the probes PVCA0063 (in vitro methylated [+Me] and unmethylated [−Me]) and PSodA, from left to right, were 0, 7.8, 15.6, 31.25, 62.5, 125, and 250 nM. The concentrations of Fur added in EMSAs with the probe PAphA, from left to right, were 0, 125, 250, and 500 nM. The positions of higher- and lower-mobility complexes are designated by HMC and LMC, respectively. Data are representative results from one of at least two independent experiments. (E) The heritability of PVCA0063 methylation was tested by taking cells grown under iron-depleted conditions (fully methylated) and incubating them for the indicated number of generations under iron-replete conditions. (F) Heterogeneity in the methylation of the VCA0063 promoter was assessed among single-colony isolates by MeSR digestion and qPCR. Each data point represents an independent biological replicate, and the horizontal line represents the median of each sample. NS, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.