Fig 1.
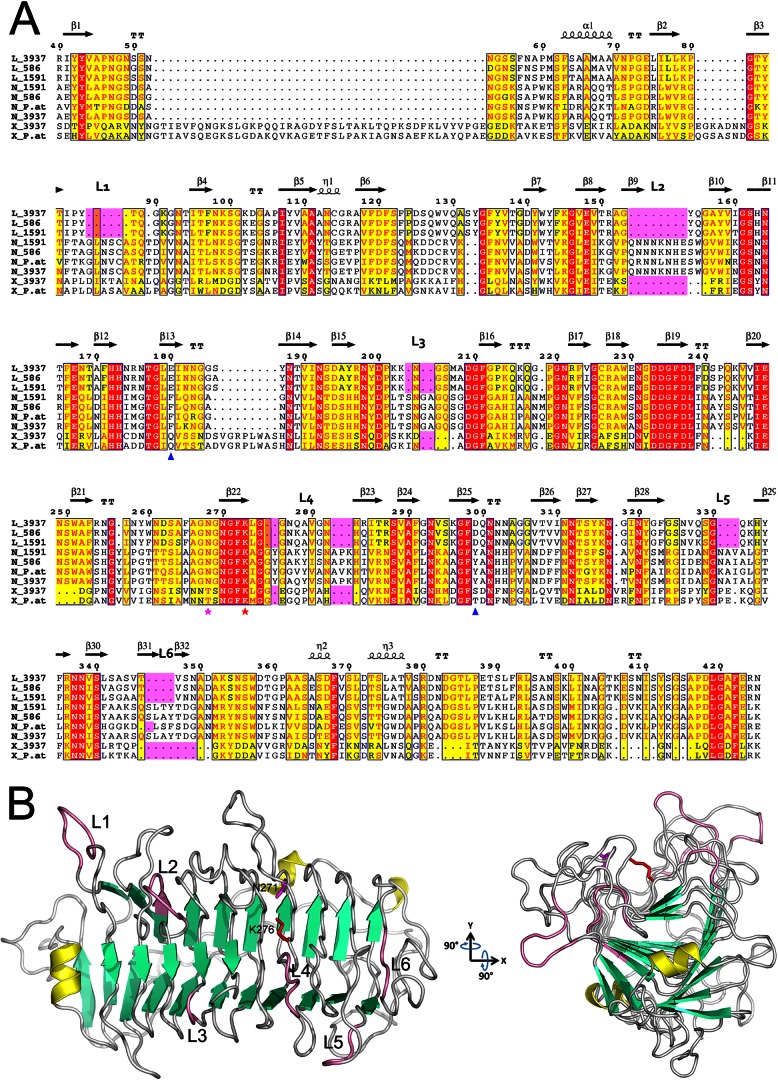
Alignment of the PL9 family pectate lyases and the PelN structural model. (A) Alignment of representative examples from the PL9 family, namely, PelL, PelN, and PelX, of Dickeya strains 3937, Ech586, and Ech1591, and of Pectobacterium atrosepticum SCRI1043 (UniProt/NCBI accession codes PLYL_DICD3, D2BWX2_DICD5, C6CF33_DICZE, C6CJG7_DICZE, D2C125_DICD5, CAG75452.1, E0SAR9_DICD3, Q9Z5P8_ERWCH, and YP_052593.1). The residue numbering is for D. dadantii 3937 PelL. Secondary structure elements identified in the crystal structure of D. dadantii PelL (PDB 1RU4) are indicated at the top. The additional loops (L1 to L6), specific for PelN, are also indicated at the top and highlighted in pink. Residues shown with red or yellow backgrounds are identical or similar, respectively, throughout the sequences. The catalytic asparagine and lysine involved in PelL activity are shown by magenta and red asterisks, respectively. Asp299 and Glu180, presumed to form calcium-mediated interactions with the substrate in PelL, are indicated with blue triangles. Alignment was generated using the ESPript web server (44). (B) The PelN model (residues 26 to 439) was generated using the program MODELLER 9v10, with the D. dadantii PelL structure (PDB 1RU4) as a template. The secondary structure elements are shown in yellow for α-helices, in green for β-strands, and in pink for specific PelN loops. Catalytic asparagine and lysine are shown in magenta and red, respectively, using the PelN numbering (N271 and K276). To highlight the presumed substrate-binding cleft and the surrounding extra loops, the structure was rotated around the x and y axes by 90°. The image in panel B was generated using PYMOL.