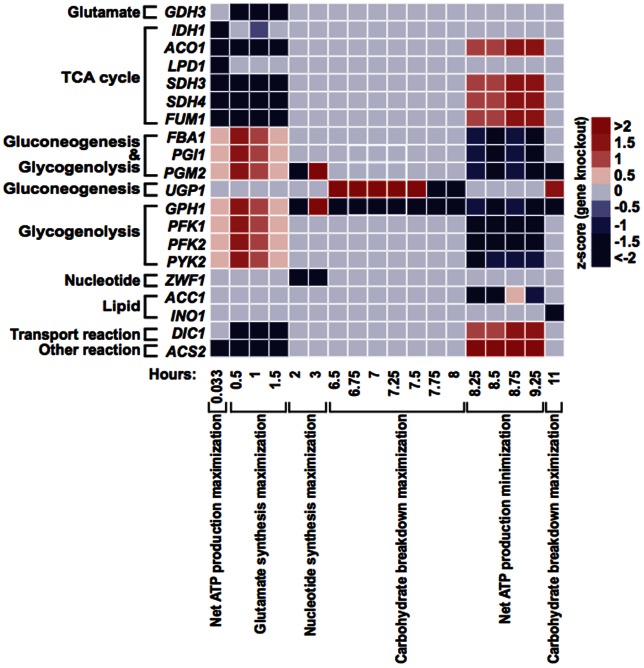
Figure 6. Model prediction of novel genes required for sporulation.
Every gene in the meiosis-specific network is deleted in silico; optimal fluxes are obtained using the best objective function combined with expression-based constraints at each time point. Pearson correlations are calculated between optimal fluxes and biochemical data on eight pathways for gene KOs. Deviation from the WT correlation is quantified by a z-score. Genes previously unknown to be required for sporulation and having a z-score ≤−2 for at least one time point are predicted to be novel sporulation-deficient genes.