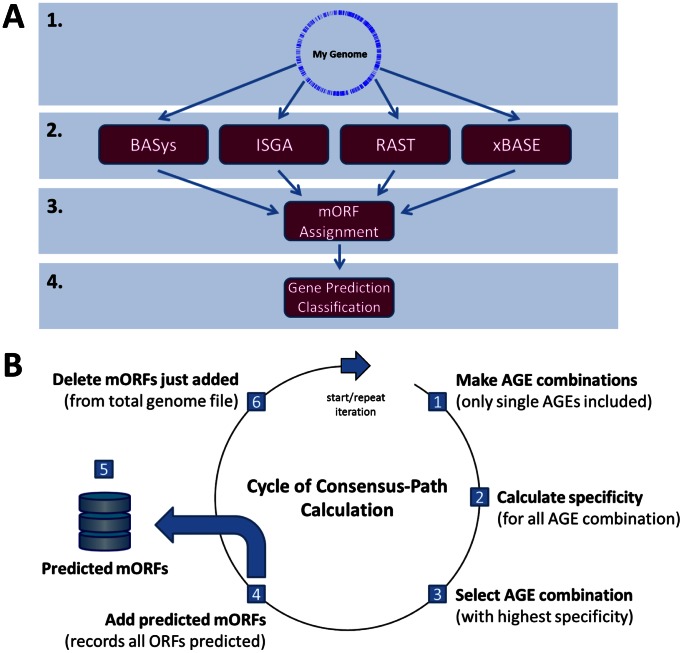
Figure 3. The methodology of comparative annotation and a step-by-step description for the consensus-path.
A. In our methodology, a single genome of interest (1) is first uploaded to the four different AGEs (2) for ORF prediction and annotation (see also Table 1). After receiving all predictions from the respective AGEs, a mORF assignment is performed (3), as described in Figure 1. Finally, on this set of mORFs for the genome of interest, the AGE gene predictions are classified (e.g., correct and incorrectly predicted mORFs) (4) as described in Fig. 2. B. With the mORFs generated as described in A, a consensus-path calculation can be performed to find the sequential (combinations of) AGEs predicting the subset of mORFs under study with the highest specificity. This cycle (or iteration) consists of the following steps: (1) generating all possible AGE combinations (single AGEs only are also included) within the set of engines used (2) calculating for each possible AGE combination (or single AGE) its specificity for that subset of mORFs considered (see formula 2 in the materials and methods). AGE combinations selected in the previous round are omitted in subsequent iterations. (3) The AGE combination (or single AGE) generating the highest specificity (hence, lowest error-rate) is selected. (4) These predicted mORFs are added to the (existing) list of predicted mORFs (5), for the genome of interest. (6) mORFs selected in (4) are removed from the mORFs file originally started with. The remaining mORFs are subjected to a next step of selecting the AGE (combination) with the highest specificity (1). This iteration is repeated until for a given genome no new mORFs are added to the prediction results in (4).