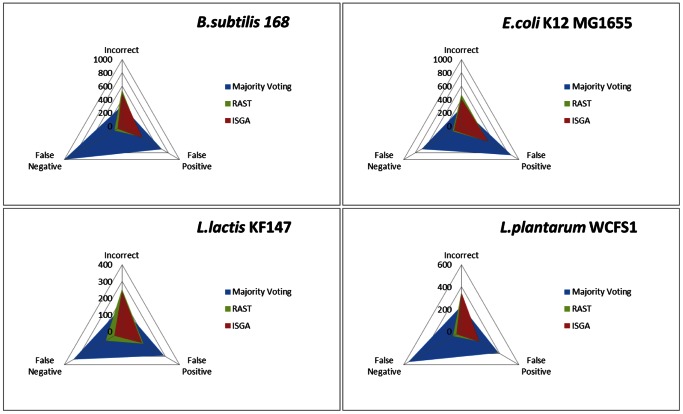
Figure 5. Start codon prediction performance by majority voting versus ISGA and RAST.
This radar-plot illustrates start codon annotation results for four moderate GC% reference genomes B. subtilis 168, E. coli K12 MG1655, L. lactis KF147 and L. plantarum WCFS1 determined by majority voting with BASys, ISGA, RAST and xBASE predictions, or by a single AGE. With majority voting a prediction is trusted if more than 50% of the AGEs predict exactly the same start codon coordinate for a given mORF. Predictions were evaluated with the reference genomes (Table 2). The axis gridlines in all directions are in steps of 100 or 200 ORFs; with incorrect predicted ORF start codons (0°), false positively (FP) and false negatively (FN) predicted ORF start codons (respectively 120° and 240°).