Abstract
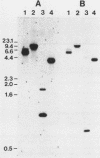
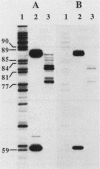
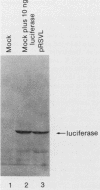
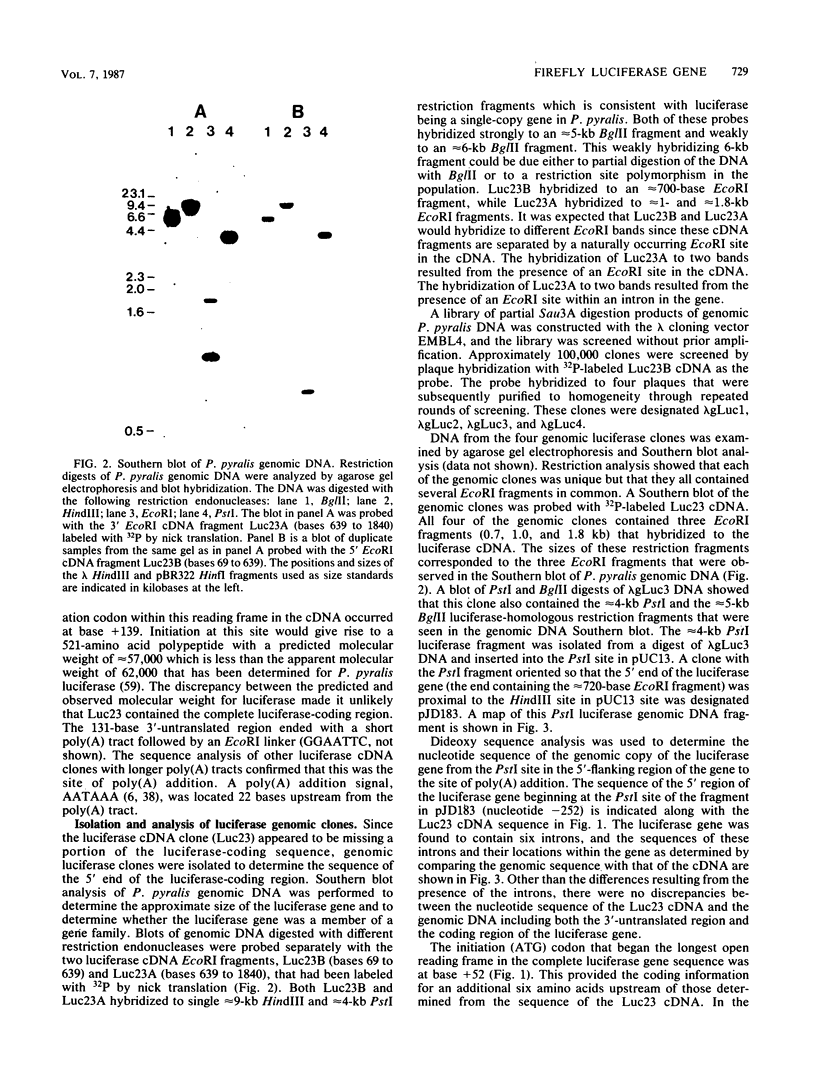
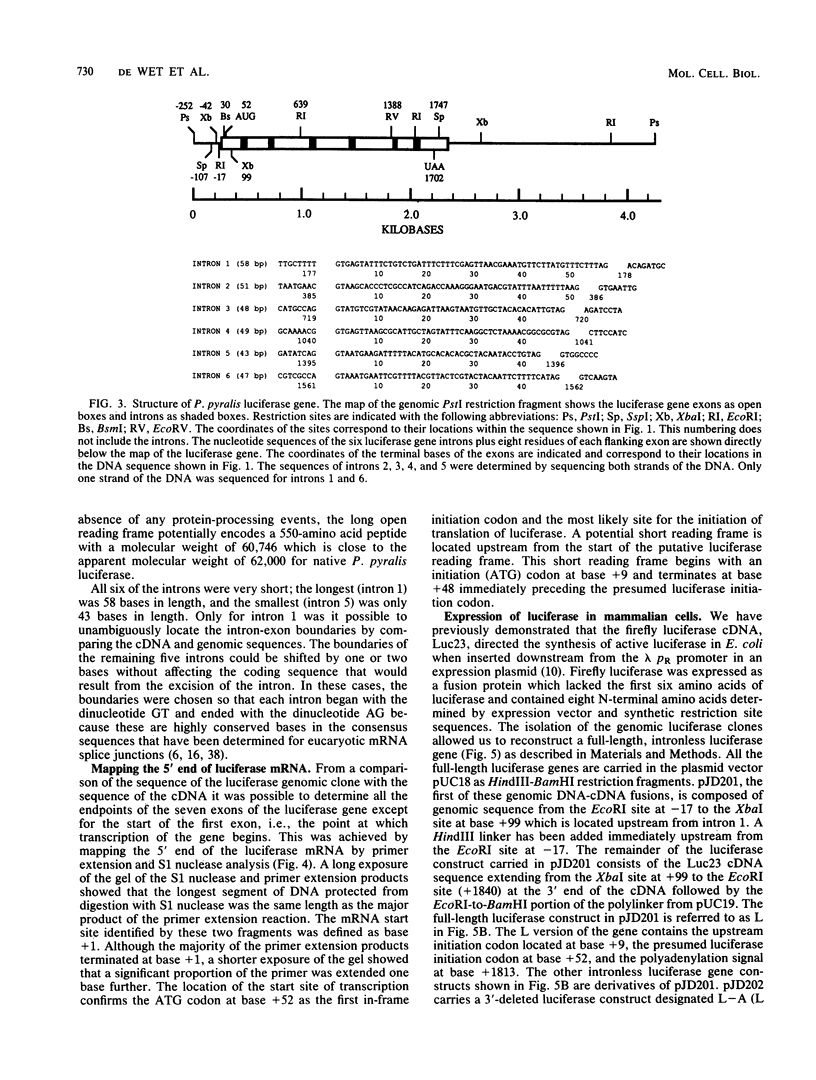
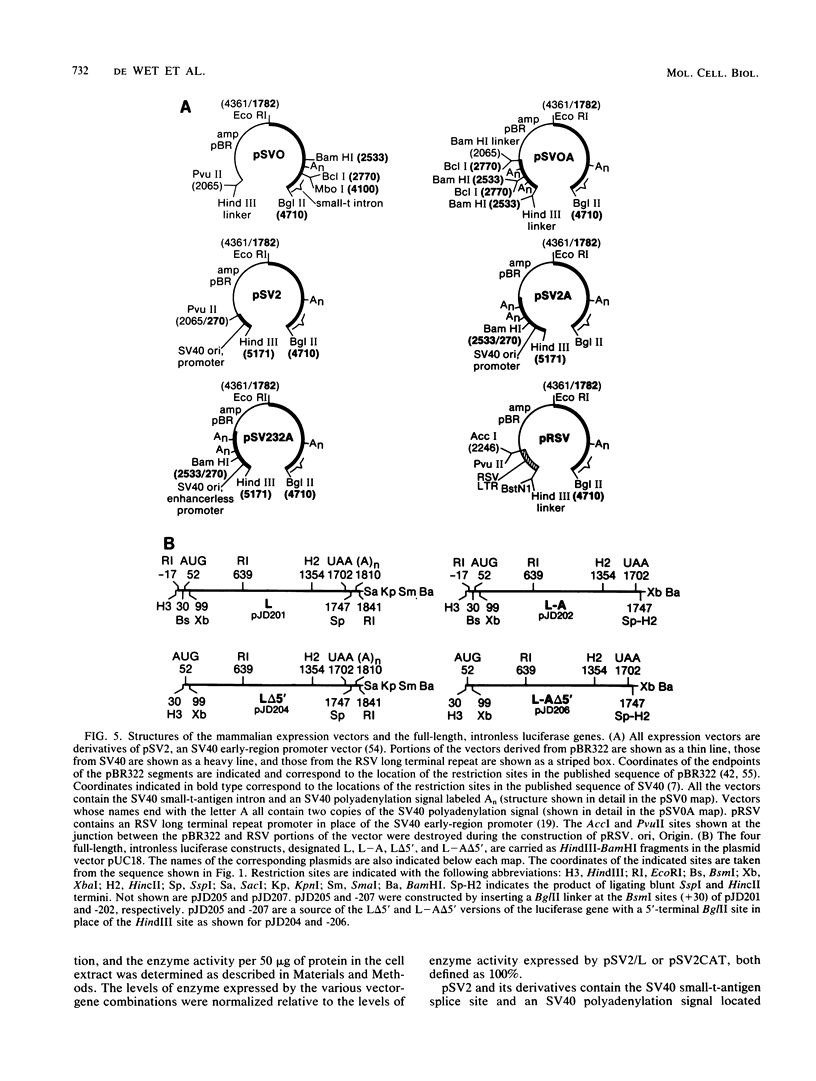
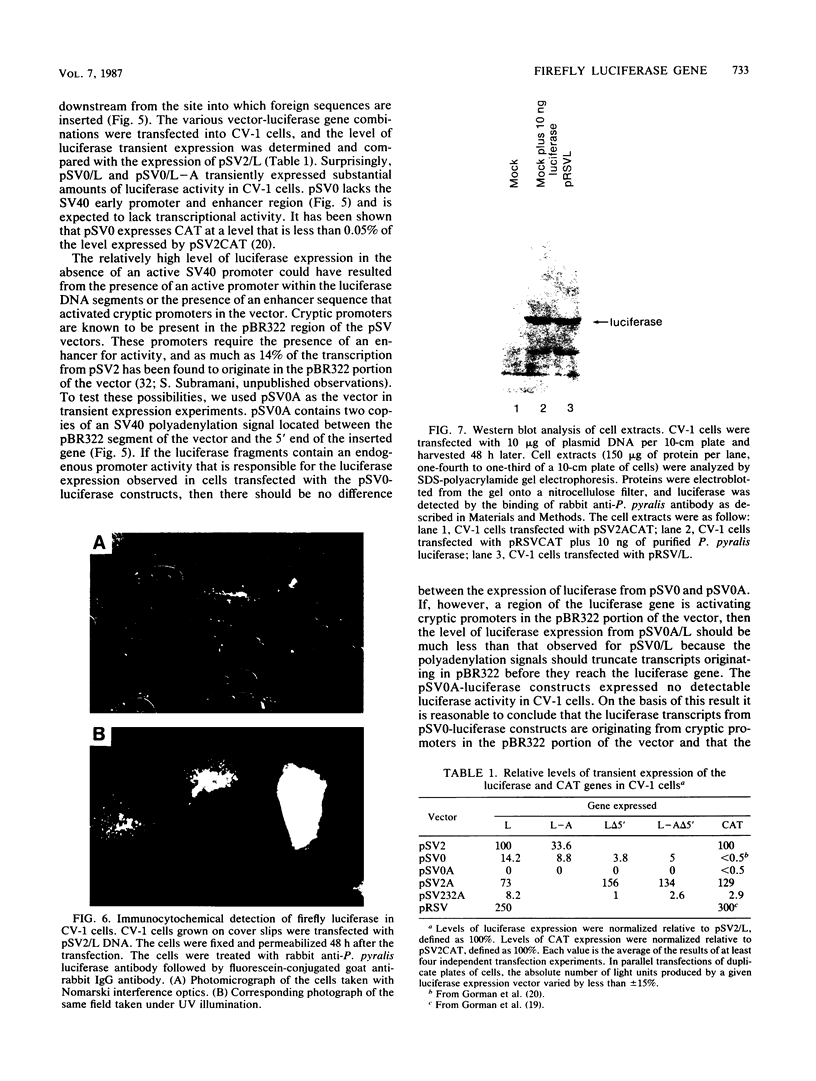
The nucleotide sequence of the luciferase gene from the firefly Photinus pyralis was determined from the analysis of cDNA and genomic clones. The gene contains six introns, all less than 60 bases in length. The 5' end of the luciferase mRNA was determined by both S1 nuclease analysis and primer extension. Although the luciferase cDNA clone lacked the six N-terminal codons of the open reading frame, we were able to reconstruct the equivalent of a full-length cDNA using the genomic clone as a source of the missing 5' sequence. The full-length, intronless luciferase gene was inserted into mammalian expression vectors and introduced into monkey (CV-1) cells in which enzymatically active firefly luciferase was transiently expressed. In addition, cell lines stably expressing firefly luciferase were isolated. Deleting a portion of the 5'-untranslated region of the luciferase gene removed an upstream initiation (AUG) codon and resulted in a twofold increase in the level of luciferase expression. The ability of the full-length luciferase gene to activate cryptic or enhancerless promoters was also greatly reduced or eliminated by this 5' deletion. Assaying the expression of luciferase provides a rapid and inexpensive method for monitoring promoter activity. Depending on the instrumentation employed to detect luciferase activity, we estimate this assay to be from 30- to 1,000-fold more sensitive than assaying chloramphenicol acetyltransferase expression.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Benyajati C., Spoerel N., Haymerle H., Ashburner M. The messenger RNA for alcohol dehydrogenase in Drosophila melanogaster differs in its 5' end in different developmental stages. Cell. 1983 May;33(1):125–133. doi: 10.1016/0092-8674(83)90341-0. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- DeLuca M., McElroy W. D. Two kinetically distinguishable ATP sites in firefly luciferase. Biochem Biophys Res Commun. 1984 Sep 17;123(2):764–770. doi: 10.1016/0006-291x(84)90295-x. [DOI] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Engebrecht J., Nealson K., Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983 Mar;32(3):773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- Engebrecht J., Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J., Simon M., Silverman M. Measuring gene expression with light. Science. 1985 Mar 15;227(4692):1345–1347. doi: 10.1126/science.2983423. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- GREEN A. A., MCELROY W. D. Crystalline firefly luciferase. Biochim Biophys Acta. 1956 Apr;20(1):170–176. doi: 10.1016/0006-3002(56)90275-x. [DOI] [PubMed] [Google Scholar]
- Garfinkel M. D., Pruitt R. E., Meyerowitz E. M. DNA sequences, gene regulation and modular protein evolution in the Drosophila 68C glue gene cluster. J Mol Biol. 1983 Aug 25;168(4):765–789. doi: 10.1016/s0022-2836(83)80074-6. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Merlino G. T., Willingham M. C., Pastan I., Howard B. H. The Rous sarcoma virus long terminal repeat is a strong promoter when introduced into a variety of eukaryotic cells by DNA-mediated transfection. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6777–6781. doi: 10.1073/pnas.79.22.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes R., Niday E., Gordon J. A dot-immunobinding assay for monoclonal and other antibodies. Anal Biochem. 1982 Jan 1;119(1):142–147. doi: 10.1016/0003-2697(82)90677-7. [DOI] [PubMed] [Google Scholar]
- Hung M. C., Wensink P. C. Sequence and structure conservation in yolk proteins and their genes. J Mol Biol. 1983 Mar 15;164(4):481–492. doi: 10.1016/0022-2836(83)90046-3. [DOI] [PubMed] [Google Scholar]
- Johansen H., Schümperli D., Rosenberg M. Affecting gene expression by altering the length and sequence of the 5' leader. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7698–7702. doi: 10.1073/pnas.81.24.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. W., Kafatos F. C. Structure, organization and evolution of developmentally regulated chorion genes in a silkmoth. Cell. 1980 Dec;22(3):855–867. doi: 10.1016/0092-8674(80)90562-0. [DOI] [PubMed] [Google Scholar]
- Kadesch T., Berg P. Effects of the position of the simian virus 40 enhancer on expression of multiple transcription units in a single plasmid. Mol Cell Biol. 1986 Jul;6(7):2593–2601. doi: 10.1128/mcb.6.7.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 1984 May 11;12(9):3873–3893. doi: 10.1093/nar/12.9.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kricka L. J., De Luca M. Effect of solvents on the catalytic activity of firefly luciferase. Arch Biochem Biophys. 1982 Sep;217(2):674–681. doi: 10.1016/0003-9861(82)90549-5. [DOI] [PubMed] [Google Scholar]
- Langner K. D., Weyer U., Doerfler W. Trans effect of the E1 region of adenoviruses on the expression of a prokaryotic gene in mammalian cells: resistance to 5' -CCGG- 3' methylation. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1598–1602. doi: 10.1073/pnas.83.6.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Nevins J. R. The pathway of eukaryotic mRNA formation. Annu Rev Biochem. 1983;52:441–466. doi: 10.1146/annurev.bi.52.070183.002301. [DOI] [PubMed] [Google Scholar]
- Parker B. A., Stark G. R. Regulation of simian virus 40 transcription: sensitive analysis of the RNA species present early in infections by virus or viral DNA. J Virol. 1979 Aug;31(2):360–369. doi: 10.1128/jvi.31.2.360-369.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody D. S., Berg P. Termination-reinitiation occurs in the translation of mammalian cell mRNAs. Mol Cell Biol. 1986 Jul;6(7):2695–2703. doi: 10.1128/mcb.6.7.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody D. S., Subramani S., Berg P. Effect of upstream reading frames on translation efficiency in simian virus 40 recombinants. Mol Cell Biol. 1986 Jul;6(7):2704–2711. doi: 10.1128/mcb.6.7.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K. W. Revised sequence of the tetracycline-resistance gene of pBR322. Gene. 1983 May-Jun;22(2-3):277–280. doi: 10.1016/0378-1119(83)90112-9. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rosenberg S. M., Stahl M. M., Kobayashi I., Stahl F. W. Improved in vitro packaging of coliphage lambda DNA: a one-strain system free from endogenous phage. Gene. 1985;38(1-3):165–175. doi: 10.1016/0378-1119(85)90215-x. [DOI] [PubMed] [Google Scholar]
- SELIGER H. H., McELROY W. D. Spectral emission and quantum yield of firefly bioluminescence. Arch Biochem Biophys. 1960 May;88:136–141. doi: 10.1016/0003-9861(60)90208-3. [DOI] [PubMed] [Google Scholar]
- Sanchez F., Tobin S. L., Rdest U., Zulauf E., McCarthy B. J. Two Drosophila actin genes in detail. Gene structure, protein structure and transcription during development. J Mol Biol. 1983 Feb 5;163(4):533–551. doi: 10.1016/0022-2836(83)90111-0. [DOI] [PubMed] [Google Scholar]
- Shaw W. V., Brodsky R. F. Characterization of chloramphenicol acetyltransferase from chloramphenicol-resistant Staphylococcus aureus. J Bacteriol. 1968 Jan;95(1):28–36. doi: 10.1128/jb.95.1.28-36.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V., Packman L. C., Burleigh B. D., Dell A., Morris H. R., Hartley B. S. Primary structure of a chloramphenicol acetyltransferase specified by R plasmids. Nature. 1979 Dec 20;282(5741):870–872. doi: 10.1038/282870a0. [DOI] [PubMed] [Google Scholar]
- Singh L., Jones K. W. The use of heparin as a simple cost-effective means of controlling background in nucleic acid hybridization procedures. Nucleic Acids Res. 1984 Jul 25;12(14):5627–5638. doi: 10.1093/nar/12.14.5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Hunkapiller M., Yuen D., Silvert D., Fristrom J., Davidson N. Cuticle protein genes of Drosophila: structure, organization and evolution of four clustered genes. Cell. 1982 Jul;29(3):1027–1040. doi: 10.1016/0092-8674(82)90466-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Subramani S., Southern P. J. Analysis of gene expression using simian virus 40 vectors. Anal Biochem. 1983 Nov;135(1):1–15. doi: 10.1016/0003-2697(83)90723-6. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Hofer E., Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984 Jul;37(3):915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- Wood K. V., de Wet J. R., Dewji N., DeLuca M. Synthesis of active firefly luciferase by in vitro translation of RNA obtained from adult lanterns. Biochem Biophys Res Commun. 1984 Oct 30;124(2):592–596. doi: 10.1016/0006-291x(84)91595-x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- de Wet J. R., Wood K. V., Helinski D. R., DeLuca M. Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7870–7873. doi: 10.1073/pnas.82.23.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]