Abstract
The localization of tail-anchored (TA) proteins, whose transmembrane domain resides at the extreme C terminus, presents major challenges to cellular protein targeting machineries. In eukaryotic cells, the highly conserved ATPase, guided entry of tail-anchored protein 3 (Get3), coordinates the delivery of TA proteins to the endoplasmic reticulum. How Get3 uses its ATPase cycle to drive this fundamental process remains unclear. Here, we establish a quantitative framework for the Get3 ATPase cycle and show that ATP specifically induces multiple conformational changes in Get3 that culminate in its ATPase activation through tetramerization. Further, upstream and downstream components actively regulate the Get3 ATPase cycle to ensure the precise timing of ATP hydrolysis in the pathway: the Get4/5 TA loading complex locks Get3 in the ATP-bound state and primes it for TA protein capture, whereas the TA substrate induces tetramerization of Get3 and activates its ATPase reaction 100-fold. Our results establish a precise model for how Get3 harnesses the energy from ATP to drive the membrane localization of TA proteins and illustrate how dimerization-activated nucleotide hydrolases regulate diverse cellular processes.
Keywords: allostery, GTPases, mechanistic enzymology, translocation
Proper localization of membrane proteins is essential for the structure and function of all cells. Tail-anchored (TA) proteins, which contain a single transmembrane domain at their extreme C terminus, comprise 3–5% of the membrane proteome (1) and mediate diverse cellular processes including protein translocation, vesicular transport, and protein quality control (1, 2). Due to their topology, TA proteins cannot engage cotranslational protein targeting machineries and instead, must use posttranslational mechanisms for efficient and accurate delivery to the target membrane (1, 3).
In the guided entry of tail-anchored protein (GET) pathway, a complex protein interaction cascade delivers TA proteins to the endoplasmic reticulum (ER) (1, 2, 4, 5). TA proteins are initially captured by the cochaperone Sgt2 in yeast (6, 7) or the BAG6 complex in mammalian cells (8). The Get4/5 complex (or its mammalian homolog TRC35/Ubl4a), which binds both Sgt2 (or Bag6) and the Get3 ATPase (or its mammalian homolog TRC40) (5, 9), then enables the loading of TA protein from Sgt2 onto Get3, the central ATPase in the pathway (6, 10). The Get3/TA complex then binds its receptor, the Get1/2 complex on the ER membrane, upon which the TA protein is released from Get3 and inserted into the membrane (5, 11, 12).
TA protein insertion is an ATP-dependent process (3) driven by Get3/TRC40, an obligate ATPase homodimer (4, 5, 13–15). Twenty-one Get3 structures, solved in various nucleotide states, show that nucleotide occupancy in the Get3 ATPase domain allows adjustments at its dimer interface that are amplified into larger displacements of its helical domains. This leads to various structures, from open conformations in apo-Get3 in which the helical subdomains are separated, to more closed conformations in adenylyl imidodiphosphate or ADP⋅AlF4–-bound Get3 in which the helical domains form a contiguous hydrophobic groove proposed to mediate TA protein binding (14–18). Further, the Get1 cytosolic domain preferentially binds apo-, open Get3 (11, 12, 19), strongly suggesting that Get1 promotes the release of nucleotide and TA proteins from Get3 at the end of the targeting cycle.
Despite rich structural information, many key questions remain regarding how the Get3 ATPase cycle drives the efficient delivery of TA proteins. First, when, where, and how ATP binding and hydrolysis occur in the GET pathway has been unclear. Second, ADP-bound Get3 has been solved in both open and closed structures (14, 18), raising questions as to the specificity of Get3 in recognizing nucleotides and generating nucleotide-driven conformational changes. Third, the nucleotide states of Get3 required for interacting with Get4/5 or for Get4/5-mediated loading of TA proteins remain controversial (6, 10, 20). Most importantly, models based on a two-state open-to-closed transition are insufficient to explain the complex cascade of protein interactions that must be coordinated by Get3, which requires multiple functional states in this ATPase.
The requirement for the Sgt2⋅Get4/5 complex in the GET pathway raises additional questions. Why is Get3 unable to directly capture the TA substrate? How does the Get4/5 complex drive the transfer of TA proteins to Get3? Thus far, Get4/5 appears to be nothing more than a scaffold that brings Sgt2 and Get3 into close proximity. Whether Get4/5 can actively facilitate TA protein capture by Get3 is unclear.
Finally, whereas the predominant model for TA protein binding invokes a closed Get3 dimer (14), there is also evidence for a tetrameric Get3 complex: recombinant Get3/TA complexes are predominantly tetramers in size exclusion chromatography, and several archaeal Get3 homologs form obligate tetramers (16, 21). Whether and how a Get3 tetramer functions in TA protein targeting remain unclear.
To address these questions, here we establish a quantitative framework for the ATPase cycle of Saccharomyces cerevisiae (Sc) Get3. We demonstrate that Get4/5 and the TA protein substrate actively regulate this cycle to ensure the precise timing of ATP hydrolysis. These results provide an explicit model for how Get3’s ATPase cycle is coupled to conformational changes that drive TA protein targeting.
Results
Cooperative ATP Binding to Get3.
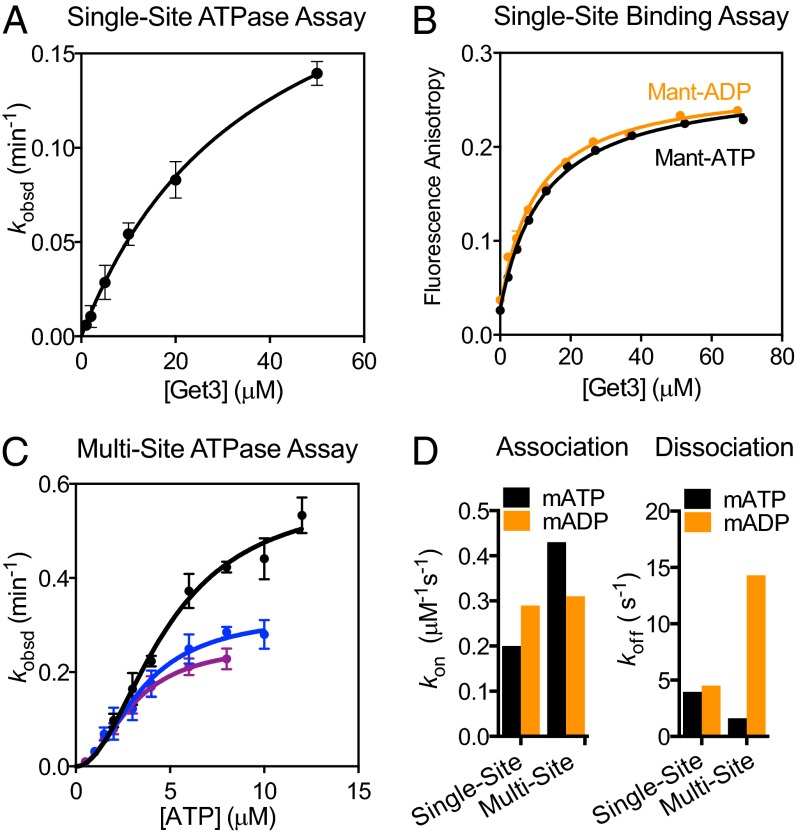
We began by establishing a quantitative framework for the Get3 ATPase cycle (Fig. 1). To probe for nucleotide-driven conformational changes, we compared Get3’s activity under two conditions: (i) “Single-site” conditions, in which Get3 is in 10- to 1,000-fold excess over the nucleotide so that statistically, the majority of nucleotide-bound Get3 dimers have a single ATPase site occupied; and (ii) “multi-site” conditions, in which the nucleotide is in excess over Get3 so that both ATPase sites are occupied. Nucleotide binding to Get3 is measured using both ATPase assays (Fig. 2A and Fig. S1A) and direct measurements based on changes in anisotropy of the fluorescent ATP analog 2′-/3′-O-(N′-methylanthraniloyl)-ATP (mantATP) (Fig. 2B). Under single-site conditions, Get3 binds ATP weakly and displays no discrimination between ATP and ADP (Fig. 1, Fig. 2 A and B, and Table S1, K1 and K9). In contrast, under multi-site conditions Get3’s ATPase reaction exhibited a cooperative dependence on ATP concentration, giving a Hill coefficient of 2 and a ∼10-fold higher affinity for binding of the second ATP (Figs. 1 and 2C and Table S1, K3).
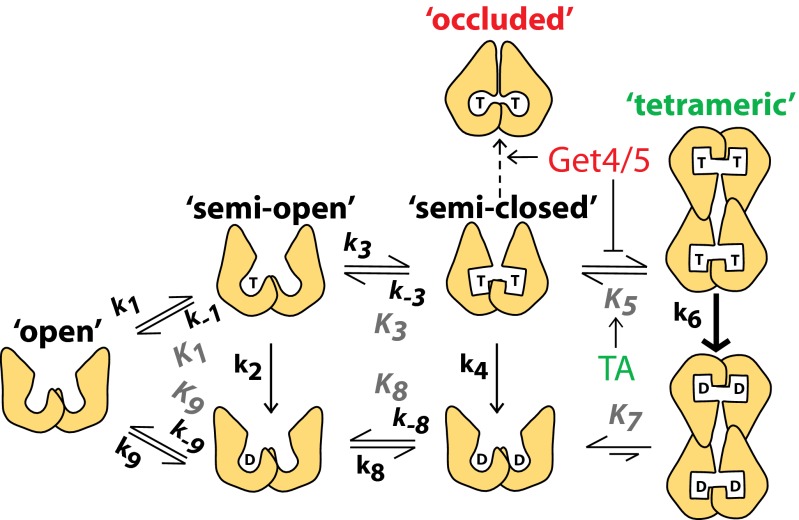
Fig. 1.
Model for the ATPase cycle of Get3. T denotes ATP; D denotes ADP. Shapes depict various Get3 conformations: steps 1 and 2, ATP binding and hydrolysis by a single active site in Get3; step 3, ATP binding to a second active site of Get3; step 4, ATP hydrolysis from dimeric Get3; step 5, formation of the Get3 tetramer; steps 6 and 7, ATP hydrolysis and ADP release from tetrameric Get3; steps 8 and 9, release of ADP from the two active sites of Get3. Individual rate and equilibrium constants are listed in Table S1.
Fig. 2.
Cooperative ATP binding to Get3. (A) Single-site ATP hydrolysis by Get3. Data were fit to Eq. S7 and they gave a KM of 37 ± 6.7 µM. (B) Equilibrium titration of mantATP (0.3 µM, black) and mantADP (0.3 µM, gold) binding to Get3 under single-site conditions. Data were fit to Eq. S1. (C) ATP hydrolysis by Get3 under multi-site conditions. Data were fit to Eq. S8 and they gave a Hill coefficient of 2, average KM values of 3.0 ± 0.2, 3.6 ± 1.0, and 4.8 ± 0.2 µM, and observed kcat values of 0.26 ± 0.02, 0.33 ± 0.03, and 0.58 ± 0.03 min−1, respectively, for reactions with 0.2 (purple), 0.5 (blue), or 1.0 (black) µM Get3. (D) Summary of nucleotide binding and release kinetics. See also Table S1.
To test the specificity of this cooperative effect, we directly measured the rates of nucleotide binding to and release from Get3 using: (i) environmentally sensitive changes in mantATP under single-site conditions (Fig. S1B), and (ii) FRET between a native tryptophan in Get3 and mantATP under multi-site conditions (Fig. S1C) (12). These measurements show that ATP binds twofold faster and dissociates threefold more slowly under multi-site conditions (Fig. 2D and Fig. S2, black; Fig. 1 and Table S1, k1, k−1 vs. k3, k−3), providing independent support for cooperative ATP binding to Get3. This cooperativity is specific for ATP: compared with single-site conditions, the rate of mantADP binding was unchanged, and ADP release is over threefold faster under multi-site conditions (Fig. 2D and Fig. S2, gold; Fig. 1 and Table S1, k8, k−8 vs. k9, k−9), indicating that Get3 disfavors ADP occupancy at both active sites. Together, these results show that ATP specifically induces rearrangements in Get3 that lead to stronger binding of the second ATP molecule (Fig. 1, steps 1 and 3), whereas ADP does not (Fig. 1, step 8).
Tetramerization of Get3 Activates ATP Hydrolysis and Is Required for TA Protein Targeting.
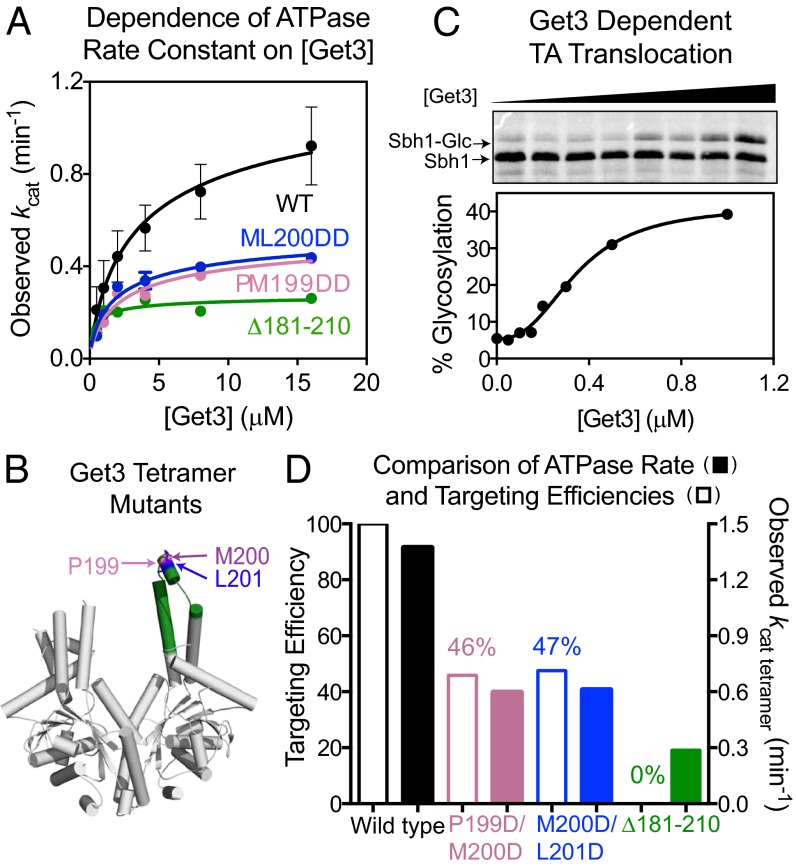
Unexpectedly, the observed ATPase rate constant at saturating ATP concentrations, or kcat, rises with increasing Get3 concentration (Figs. 2C and 3A). This phenomenon was observed even in the presence of BSA, an effective surfactant and crowding reagent, suggesting that it is unlikely to be caused by enzyme loss or inactivation at low concentrations (Fig. S3A). Instead, this result suggests that an oligomerization process stimulates Get3’s ATPase activity. Quantitatively, these data are most consistent with a model in which dimeric Get3 is in dynamic equilibrium (Kd = 3.5 ± 1.9 µM) with tetrameric Get3, which hydrolyzes ATP faster than dimeric Get3 (Fig. 1, steps 5–7 and Eq. S9). Analysis of the data based on this model yielded a kcat value for tetrameric Get3 of 1.3 ± 0.4 min−1 (Fig. 1 and Table S1, k6), over 100-fold faster than dimeric Get3 (Fig. 3A and Fig. S3A; Fig. 1 and Table S1, k4). This phenomenon has previously escaped detection, likely because it is abolished in less physiological solution conditions (Fig. S3B), whereas our ATPase measurements used the same buffer as for protein targeting/translocation reactions (22). The transient nature of tetrameric Get3 could also render it susceptible to dissociation during size exclusion chromatography (23).
Fig. 3.
Tetramerization stimulates Get3’s ATPase activity and is required for TA protein targeting. (A) Observed kcat values as a function of Get3 concentration, for wild-type Get3 (black) and mutants Δ181–210 (green), P199D/M200D (pink), and M200D/L201D (blue). Data were fit to Eq. S9 and summarized in D. (B) Structure of ScGet3 (Protein Data Bank: 3A36) highlighting the residues mutated. The remainder of residues 181–210 is in green. (C) Targeting and insertion of Sbh1p by wild-type Get3. Data were fit to Eq. S11 and they gave a Hill coefficient of 2. (D) Comparison of TA targeting efficiencies (open) and tetramer ATPase rate constants (filled) for wild-type Get3 (black) and mutants P199D/M200D (pink), M200D/L201D (blue), and Δ181–210 (green). Percent translocation was normalized to wild-type Get3.
In a structure of the Methanocaldococcus jannaschii (Mj) Get3 tetramer, helix 8 plays a key role in stabilizing the tetramer interface. Mutations of conserved hydrophobic residues in this helix, F192D, M193D, and M196D, destabilize the tetramer (21). To independently test whether tetramerization of ScGet3 is responsible for ATPase activation, we mutated homologous residues in ScGet3 (P199D/M200D, M200D/L201D; Fig. 3B). Given their location, these mutations are unlikely to affect the TA binding groove of the dimer, but would specifically disfavor the formation or conformation of the tetramer. These mutations reduced activated ATP hydrolysis at high Get3 concentrations to almost the same extent as mutant ∆181–210, a negative control that lacks a large portion of the putative TA-binding groove (Fig. 3 A and B) and completely abolished TA protein capture and targeting (Fig. S4D). In contrast, the kcat values at low Get3 concentrations, where it is primarily a dimer, were largely unchanged in these mutants (Fig. 3A). As additional controls, we mutated residues in the putative TA binding groove of ScGet3 in the dimer model (F190D or I193D) (14). In contrast to the mutants designed to disrupt the tetramer, F190D and I193D exhibit over 10-fold higher ATPase activity and tetramerize more favorably than wild-type Get3 (Fig. S3C and Table S2). These results provide independent evidence that formation of a Get3 tetramer is required for activated ATP hydrolysis.
If tetramerization of Get3 and its associated ATPase activation were important, it would also be manifested in a targeting reaction. To test this hypothesis, we quantitatively measured the targeting and translocation of a TA substrate, Sbh1p, to ER microsomes (Fig. S4A). An NXT glycosylation site was engineered into the C terminus of Sbh1p, whose glycosylation reports on successful translocation across the membrane. Both the translation lysate and ER microsomes were derived from a Δget3 strain, so that Sbh1p targeting is dependent solely on exogenously added Get3. The efficiency of Sbh1p targeting and translocation exhibited a cooperative dependence on Get3 concentration with a Hill coefficient of 2 (Fig. 3C and Fig. S4B), suggesting that efficient targeting requires two Get3 dimers to further associate to form a tetramer. Additionally, mutants P199D/M200D and M200D/L201D exhibit defects in targeting (Fig. 3D and Fig. S4C) that quantitatively correlate with their defects in tetramerization-induced ATPase activation (Fig. 3D). Combined with previous observations that mutants M200D and L201D are deficient in TA substrate binding and supporting cell growth (14), these results provide strong evidence that transient formation of a Get3 tetramer is required for efficient TA protein targeting.
Get4/5 Enhances ATP Binding but Inhibits ATP Hydrolysis by Get3.
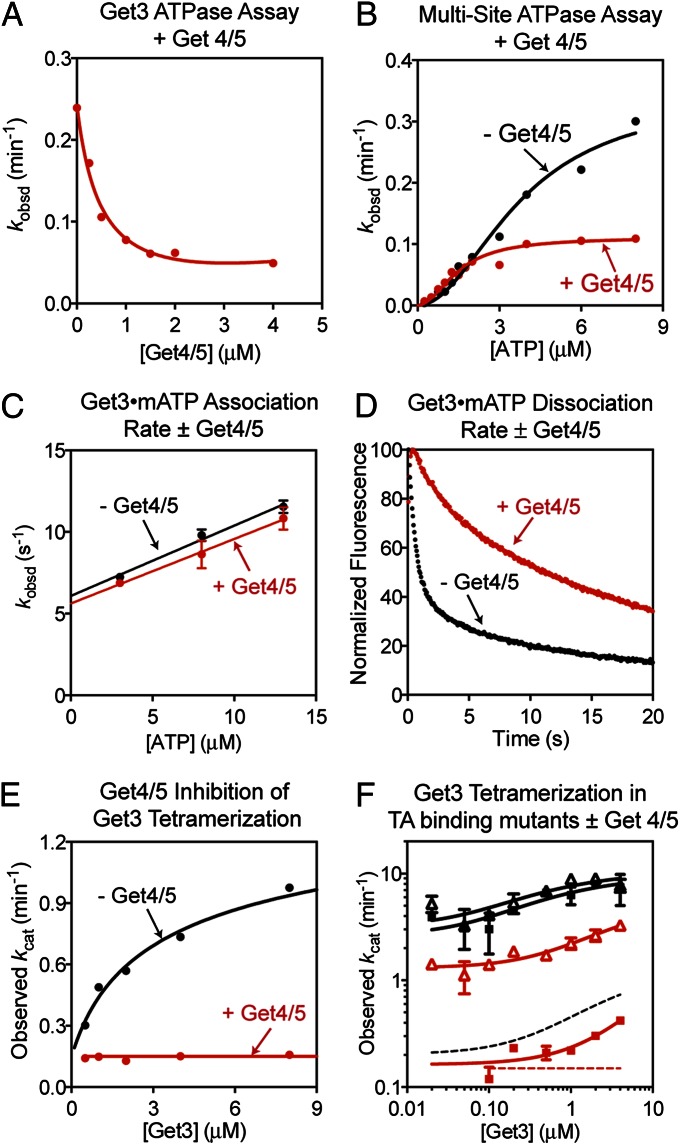
We next asked how the Get4/5 complex, which acts as a scaffold to facilitate TA protein loading from Sgt2 onto Get3, regulates the Get3 ATPase. Intriguingly, Get4/5 stoichometrically inhibits the ATPase activity of Get3 (Fig. 4A and Fig. S5A). Analysis of the ATP concentration dependence of the reaction showed that the average KM value is lowered to 1.4 ± 0.3 µM with Get4/5 present, indicating that Get3 binds ATP more strongly when it is bound to Get4/5 (Fig. 4B and Fig. S5B). In contrast, Get4/5 reduced the value of kcat, indicating specific inhibition of ATP hydrolysis (Fig. 4B). Thus, Get4/5 induces Get3 into an alternative conformation in which ATP is bound more tightly but held in a catalytically compromised structure.
Fig. 4.
Get4/5 tightens ATP binding to Get3 and inhibits its ATPase activity. (A) Get4/5 stoichiometrically inhibits Get3’s ATPase activity. Reaction contained 0.5 µM Get3 and 10 µM ATP. (B) ATP concentration dependence of ATPase activity at 0.5 µM Get3, with (red) or without (black) 5 µM Get4/5 present. Data were fit to Eq. S8 and they gave average KM values of 3.7 ± 0.2 and 1.4 ± 0.3 µM with and without Get4/5, respectively. (C) Kinetics of mantATP binding to Get3 with (red) and without (black) 3.0 µM Get4/5 present. Data were fit to Eq. S4. (D) Dissociation of mantATP from Get3 was slowed in the presence (red) of 3.0 µM Get4/5. (E) Observed kcat values as a function of Get3 concentration with (red) or without (black) 50 µM Get4/5 present. Data with Get3 were analyzed as in Fig. 3A, and data with the Get3⋅Get4/5 complex were fit to a linear function. (F) Same as E but with Get3 mutants F190D (triangles) and I193D (squares) with (red) or without (black) Get4/5 present. Dotted lines are fits for wild-type Get3 in E and shown for comparison. All rate constants are reported in Tables S2 and S3.
To provide independent evidence for this model, we tested how Get4/5 alters nucleotide binding of Get3 using the FRET assay. Get4/5 did not affect the rate of ATP binding to Get3 (Fig. 4C) but reduced the rate of ATP dissociation from Get3 at least 10-fold (Fig. 4D), providing direct evidence that Get3 binds ATP more tightly when it is bound to Get4/5. This effect is specific for ATP, as under the same conditions ADP release from Get3 remained fast and was largely unaffected by Get4/5 (Fig. S5C).
If Get4/5 induces stronger ATP binding to Get3, then ATP-bound Get3 would also bind more strongly to Get4/5. To test this prediction, we measured complex formation between Get3 and Get4/5 using gel filtration chromatography. With apo-Get3, complex assembly was not detected even at micromolar protein concentrations (Fig. S5D). In contrast, with saturating ATP present, almost all Get3 formed a complex with Get4/5 (Fig. S5E). These results, although qualitative, are consistent with previous pull-down experiments in which a stable Get3-4/5 complex was enriched in the presence of nucleotides (6, 10, 20). Together, these results show that Get4/5 preferentially binds ATP-loaded Get3 and reciprocally, interaction with Get4/5 enables ATP to be more tightly bound to Get3.
As the Get3 ATPase activity is activated upon tetramerization, we asked whether Get4/5 inhibits this activation. Get4/5 also stoichiometically inhibits the ATPase reaction at high Get3 concentrations, where it is predominantly a tetramer (Fig. S5A). With saturating Get4/5 and ATP, the ATPase rate constant stayed constant at 0.16 ± 0.07 min−1 and was independent of Get3 concentration (Fig. 4E). Thus, Get4/5 inhibits formation of the Get3 tetramer or the ATPase activation induced by tetramerization.
Mutants F190D and I193D exhibit higher ATPase activities than wild-type Get3 in both the dimeric and tetrameric forms; both of these activities are substantially reduced in the presence of Get4/5 (Fig. 4F). Thus, these superactive mutant ATPases provide stronger evidence that the ATPase activity of dimeric Get3 is also inhibited by the Get4/5 complex.
TA Protein Induces Rapid ATP Hydrolysis and Locks Get3 in the ADP-Bound State.
We next asked how the TA protein substrate regulates the Get3 ATPase. To this end, we coexpressed Get3 with Sbh1p. The Get3/Sbh1 complex purified predominantly as a tetrameric complex (Fig. S6A), consistent with previous observations (16, 21).
To determine the ATP hydrolysis rate from this complex, we carried out presteady-state measurements using a high ATP concentration and Get3 active sites in 1:5 stoichiometry relative to ATP. Under these conditions, the ATPase reaction exhibited two distinct kinetic phases: (i) an initial burst whose magnitude increased with increasing Get3 concentration (Fig. 5A and Fig. S6B), representing a rapid first round of ATP hydrolysis; and (ii) a slower linear phase representing subsequent rounds of ATP turnover at steady state. The rate constant for the first round of ATP hydrolysis is 3.3 ± 1.1 min−1 (Eq. S10), over 100-fold faster than that of the Get3 dimer. The rate constant for steady-state ATP turnover is 0.055 ± 0.001 min−1, 60-fold slower than the first turnover. Thus, loading of TA protein onto Get3 activates one round of ATP hydrolysis, but subsequent ATP turnover is inhibited. Further, ATPase activation in the Get3/TA complex was not observed under single-site conditions (compare Figs. 5B and 2A), suggesting that it requires both Get3 active sites to be bound with ATP. Finally, the magnitude of the burst phase is stoichiometric with the concentration of Get3 active sites, suggesting that all four ATPs in the Get3 tetramer are hydrolyzed during the first turnover.
Fig. 5.
TA substrate induces rapid ATP hydrolysis. (A) Presteady-state ATPase reaction at a 1:5 ratio of Get3/TA:ATP. The data were fit to Eq. S10. (B) ATP hydrolysis from the Get3/TA complex under single-turnover conditions. Data were fit to Eq. S7 and they gave a kcat value of 0.42 min−1 and a KM value of 33 µM. (C) Kinetics of mantATP binding to the Get3/TA complex. Two phases were observed. The dashed part of the curve depicts theoretical increases in binding rates at lower ATP concentrations where bimolecular association is rate limiting, but which was inaccessible in our experiments. (D) MantADP dissociation from the Get3/TA complex. Data with Get3 (black) were from Fig. S2F (black) and are shown for comparison. All rate constants are reported in Table S3.
To test whether nucleotide binding or release could be rate limiting for the observed ATPase rates, we used the fluorescence assays to directly measure these events. MantATP binding to the Get3/Sbh1 complex was slow and concentration independent at the lowest concentrations tested under both multi-site (Fig. 5C and Fig. S6C) and single-site conditions (Fig. S6D), suggesting that a slow conformational change of the Get3/Sbh1 complex becomes rate limiting for ATP binding. The rate of the dominant, slow phase in ATP binding is similar to that of the burst phase in the ATPase reaction (5.0 vs. 3.3 min−1), suggesting that the ATPase rate constant observed here may still be limited by a conformational change that precedes hydrolysis.
Remarkably, dissociation of ADP is at least 100-fold slower in the Get3/Sbh1 complex compared with free Get3 (Fig. 5D and Table S3) and is indistinguishable from that of ATP or nonhydrolyzable ATP analogs (Fig. S6E and Table S3), suggesting that the nucleotides are bound tightly and shielded from solvent in this complex. Nevertheless, ADP release from the Get3/TA complex is still 200-fold faster than the steady-state ATPase rate and is unaffected by up to 10 mM inorganic phosphate (Fig. S6E and Table S3). This indicates that an additional conformational step, rather than product release, is rate limiting for steady-state ATP turnover. Together, these data argue that only one round of ATP hydrolysis occurs in the GET pathway, after which the Get3/TA complex is locked in a catalytically inactive state loaded with ADP, and disassembly of this complex would be needed to reset its ATPase cycle.
Discussion
Efficient and accurate delivery of membrane proteins often requires energy input from nucleotide triphosphates, which in the GET pathway is harnessed and used by the Get3 ATPase (13, 24). When, where, and how ATP binding and hydrolysis occur in the GET pathway remain open questions. Little is known about how Get3’s nucleotide state, conformation, and activity are regulated during TA protein targeting. Here, quantitative mechanistic analyses define a precise framework for Get3’s ATPase cycle and elucidate how it is used to drive this fundamental cellular process.
Previous work showed that Get3’s ATPase domain acts as a fulcrum at the dimer interface to generate a variety of structures (13). The cooperative ATP binding observed here supports a model in which Get3 changes from a largely open conformation in apo-Get3 to increasingly closed conformations upon successive ATP binding (Fig. 1, steps 1 and 3). Importantly, this cooperativity is specific to ATP but not ADP. Thus, an ADP-bound Get3 dimer remains in a largely open conformation (15, 18), despite the occasional observation of “closed”, ADP-bound Get3 structures (16). Nevertheless, the cooperativity induced by ATP is fairly modest, ∼10-fold. Together with previous work (25), we speculate that Get3 exists in an ensemble of conformations that are in close equilibrium with one another, and each ATP binding event induces a modest shift in the conformational landscape. Thus, even the Get3 dimer bound with both ATPs is not completely closed, and is termed semiclosed here (Fig. 1).
Intriguingly, Get3 is catalytically activated through tetramerization (Fig. 1, steps 5 and 6). This phenomenon was previously suggested by the structure of an MjGet3 tetramer and by the formation of tetrameric Get3/TA complexes (21). Our findings provide a function for tetrameric Get3, showing that it is the active species for ATP hydrolysis and for efficient TA protein targeting. In further support of this model, hydrophobic residues in helix 8 that stabilize the tetramer interface are conserved (15, 21); their mutations disrupt ATPase activation and protein targeting by Get3 (this work) and lead to defects in cell viability and TA binding (14, 21). Given the location of these residues, these phenotypes are difficult to reconcile with a dimeric model for Get3. Although each of these observations can be explained by alternative models, activation of Get3 via tetramerization provides a cohesive, unifying model that explains this diverse collection of data.
In vivo, tetramerization of Get3 by itself should be disfavored to minimize futile ATPase cycles. This could be achieved in part by the low in vivo concentration of Get3, ∼1 μM (26), which is below the Kd value for tetramerization (3.5 μM). The results here further show that futile ATPase cycles of Get3 are minimized by the Get4/5 complex, which mediates the loading of TA proteins from Sgt2 onto Get3 (6, 10). Despite previous reports of Get4/5 binding to apo-Get3 (27), our results demonstrate that Get4/5 preferentially binds ATP-loaded Get3 and locks it in the ATP-bound state (Fig. 6, step 2). This is achieved by tightening Get3’s ATP binding but inhibiting its hydrolytic activity, particularly the tetramerization-induced activation of ATP hydrolysis. Get4/5 could exert these effects by inducing Get3 into a distinct, “occluded” conformation in which its ATPase site is more closed but incompetent for hydrolysis (Fig. 1). In addition, Get4/5 could prevent Get3’s tetramerization. The latter model is particularly attractive as it explains why Get5 is a stable dimer (28): a complete Get4/5 complex could hold two closed Get3 dimers in the ATP-bound state, priming them for subsequent tetramer formation once the TA protein is loaded onto Get3 (Fig. 6, step 3). Regardless of the model, our data show that Get4/5 is not a passive scaffold that simply brings Sgt2 and Get3 into close proximity. Rather, Get4/5 actively promotes TA protein loading onto Get3 by locking it in the correct nucleotide state and priming its conformation for TA substrate capture (6, 8).
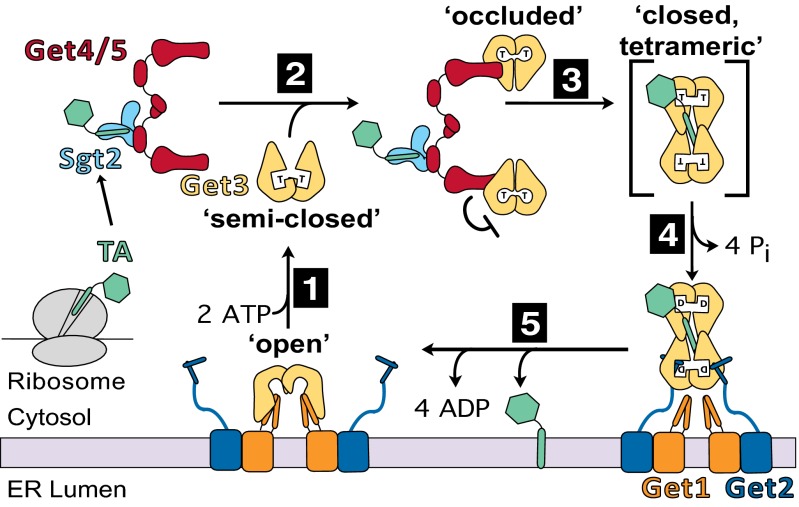
Fig. 6.
Model for TA protein targeting driven by the ATPase cycle of Get3, as described in the text.
In contrast to Get4/5, multiple lines of evidence strongly suggest that the TA protein induces the tetramerization and activation of Get3’s ATPase activity (Fig. 6, step 3): (i) coexpression of TA protein with Get3 results in a stable Get3 tetramer (this work and refs. 16, 21); and (ii) rapid ATP hydrolysis was observed with the Get3/TA complex, as would be expected for an activated Get3 tetramer. Several important lessons are learned from analysis of the Get3/TA complex. First, after the first round of ATP hydrolysis, subsequent ATP turnover is 60-fold slower and incompatible with the time scale of protein targeting in vivo, arguing that only one round of ATP hydrolysis occurs in the GET pathway. Second, following ATP hydrolysis, Get3 is locked in a catalytically inactive state. Together with observations with the Get3⋅Get4/5 complex, these results demonstrate that the open-to-closed rearrangement of Get3 can be conceptually and experimentally uncoupled: even when Get3 is globally closed and nucleotide release is slow, additional active site adjustments specifically regulate catalytic activity. We speculate that this relates to local rearrangements of the switch II loops (13), which provide multiple essential catalytic residues. The ADP-bound MjGet3 tetramer structure possibly provides a view of a closed but catalytically inactive Get3 tetramer, in which the switch II loop is pulled away and incompatible for hydrolysis (21). Finally, ADP release is substantially slowed in the Get3/TA complex and becomes indistinguishable from that of ATP, suggesting that the TA protein is dominant in inducing a closed Get3 tetramer.
In the context of the targeting cycle, TA-induced Get3 tetramer formation would be beneficial as the hydrophobic transmembrane domain of the TA substrate can be completely protected in a cage at the tetrameric interface (21), minimizing its potential aggregation (Fig. 6). Our results also suggest that following hydrolysis, ADP release from the Get3/TA complex may be delayed until Get3 finds the Get1/2 membrane receptor. Tetramer disassembly by this receptor would be needed to release the TA protein. As ATP and Get1 binding to Get3 are strongly antagonistic with one another (10–12), ATP hydrolysis in the Get3/TA complex likely primes it for disassembly at the membrane.
Collectively, our results define a precise model for how the Get3 ATPase cycle is used to drive TA protein targeting (Fig. 6). Under cellular conditions, the majority of Get3 cooperatively binds ATP at both active sites, which induces it into a semiclosed conformation (step 1). ATP-loaded Get3 is preferentially captured by Get4/5, which brings Get3 into the vicinity of Sgt2 and induces the Get3 dimer into an occluded conformation in which it is further closed but ATP hydrolysis is delayed (step 2). In this configuration, Get3 is primed to capture the TA substrate from Sgt2 (step 2). Loading of TA protein induces tetramerization of Get3 (step 3), which might also drive dissociation of Get3 from Get4/5. The tetrameric Get3/TA complex undergoes a rapid round of ATP hydrolysis, giving a stable ADP-loaded complex that binds its receptor, Get1/2, at the ER membrane (step 4). Tetramer disassembly, ADP dissociation, and TA protein release into the membrane are likely coupled, resulting in Get1 bound to apo-Get3 in the open conformation (step 5). ATP binding then releases Get3 from Get1 (10–12) to reinitiate the cycle.
Get3 is the only eukaryotic ATPase in the SIMIBI [for signal recognition particle (SRP), MinD, and BioD] family of deviant P-loop NTPases, including the SRP and SRP receptor (SR) that mediate cotranslational protein targeting (29). Although the details of each system differ, the results here reveal many similarities in the regulatory principles between Get3 and SRP/SR. Both exhibit low nucleotide affinity and forego the need of external exchange factors and activating proteins as regulatory elements (30). Instead, both use dimeric complexes as the functional unit. As dimers, both undergo conformational changes on the global (open-to-closed transitions) and local (catalytic loop adjustments) scale to generate multiple functional states during an NTPase cycle. For both, these rearrangements provide key regulatory points to sense and respond to upstream and downstream components and effect the precise timing of nucleotide hydrolysis in the pathway: GTP hydrolysis in the SRP/SR complex is stalled by the translation ribosome and reactivated by the SecYEG machinery (31, 32), whereas ATP hydrolysis in Get3 is stalled by Get4/5 and activated by the TA substrate. Based on regulatory principles, Get3 could be placed in the class of NTPases regulated by dimerization (33) whose members, aside from SRP and SR, also include the human GBP1, the septins, HypB, MnmE, and the dynamin family of GTPases (33, 34). Investigation of Get3 undoubtedly enhances our understanding of the mechanism, regulation, and evolution of this ubiquitous class of regulators.
Materials and Methods
Protein Expression and Purification.
Mutant Get3s were generated using Quikchange Mutagenesis protocol (Stratagene). Wild-type and mutant Get3s were expressed and purified as described (15, 20). Purification of the Get4/5 and Get3/Sbh1 complexes is described in SI Materials and Methods.
Fluorescence Measurements.
All fluorescent nucleotides were from Jena Biosciences. All measurements were carried out at 25 °C in Get3 assay buffer (50 mM Hepes pH 7.4, 150 mM potassium acetate, 5 mM magnesium acetate, 1 mM DTT, and 10% glycerol) using a Fluorolog-3–22 spectrofluorometer (Jobin Yvon) or a Kintek stopped-flow apparatus. Determination of individual rate and equilibrium constants is described in SI Materials and Methods and Fig. S7.
ATPase Assays.
All reactions were performed in Get3 assay buffer at 25 °C with [γ-32P]-ATP (MP Biomedicals). Reactions at Get3 concentrations below 0.5 µM also included 1 mg/mL BSA. Reactions were quenched in 0.75 M potassium phosphate (pH 3.3), analyzed by polyethylenimine cellulose TLC in 1 M formic acid/ 0.5 M LiCl, and quantified by autoradiography. Observed rate constants were obtained as described (35). Determination of individual rate and equilibrium constants is described in SI Materials and Methods.
TA Protein Targeting and Translocation.
Yeast translation extracts were prepared as described (21, 36), except that an additional centrifugation step (SW55Ti, 30 min at 291,000 × g) was included before chromatography on the G25 column. Yeast microsomes were prepared as described (5, 37). Translation and translocation of TA protein is detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank J. Chartron, H. Gristick, and C. J. M. Suloway for expression constructs, purification protocols, and critical discussions; M. Sachs and C. Wu for help with yeast translation extracts; R. Schekman for help with yeast microsomes; and D. C. Rees and members of the S.-o.S. and W.M.C. groups for helpful comments. This work was supported by career awards from the David and Lucile Packard Foundation and the Henry Dreyfus Foundation (to S.-o.S.), National Science Foundation Graduate Research Fellowship DGE-1144469 (to M.E.R.), National Institutes of Health (NIH) Training Grant 5T32GM007616-33 (to M.R.), and NIH Grant R01 GM097572 (to W.M.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222054110/-/DCSupplemental.
References
- 1.Hegde RS, Keenan RJ. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2011;12(12):787–798. doi: 10.1038/nrm3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Claessen JHL, Mueller B, Spooner E, Pivorunas VL, Ploegh HL. The transmembrane segment of a tail-anchored protein determines its degradative fate through dislocation from the endoplasmic reticulum. J Biol Chem. 2010;285(27):20732–20739. doi: 10.1074/jbc.M110.120766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport TA. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995;14(2):217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128(6):1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 5.Schuldiner M, et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134(4):634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang F, Brown EC, Mak G, Zhuang J, Denic V. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell. 2010;40(1):159–171. doi: 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chartron JW, Gonzalez GM, Clemons WM., Jr A structural model of the Sgt2 protein and its interactions with chaperones and the Get4/Get5 complex. J Biol Chem. 2011;286(39):34325–34334. doi: 10.1074/jbc.M111.277798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mariappan M, et al. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466(7310):1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jonikas MC, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323(5922):1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang F, Whynot A, Tung M, Denic V. The mechanism of tail-anchored protein insertion into the ER membrane. Mol Cell. 2011;43(5):738–750. doi: 10.1016/j.molcel.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefer S, et al. Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex. Science. 2011;333(6043):758–762. doi: 10.1126/science.1207125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mariappan M, et al. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature. 2011;477(7362):61–66. doi: 10.1038/nature10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chartron JW, Clemons WM, Jr, Suloway CJM. The complex process of GETting tail-anchored membrane proteins to the ER. Curr Opin Struct Biol. 2012;22(2):217–224. doi: 10.1016/j.sbi.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mateja A, et al. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461(7262):361–366. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suloway CJM, Chartron JW, Zaslaver M, Clemons WM., Jr Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc Natl Acad Sci USA. 2009;106(35):14849–14854. doi: 10.1073/pnas.0907522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bozkurt G, et al. Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc Natl Acad Sci USA. 2009;106(50):21131–21136. doi: 10.1073/pnas.0910223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J, Li J, Qian X, Denic V, Sha B. The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion. PLoS ONE. 2009;4(11):e8061. doi: 10.1371/journal.pone.0008061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamagata A, et al. Structural insight into the membrane insertion of tail-anchored proteins by Get3. Genes Cells. 2010;15(1):29–41. doi: 10.1111/j.1365-2443.2009.01362.x. [DOI] [PubMed] [Google Scholar]
- 19.Kubota K, Yamagata A, Sato Y, Goto-Ito S, Fukai S. Get1 stabilizes an open dimer conformation of get3 ATPase by binding two distinct interfaces. J Mol Biol. 2012;422(3):366–375. doi: 10.1016/j.jmb.2012.05.045. [DOI] [PubMed] [Google Scholar]
- 20.Chartron JW, Suloway CJM, Zaslaver M, Clemons WM., Jr Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc Natl Acad Sci USA. 2010;107(27):12127–12132. doi: 10.1073/pnas.1006036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suloway CJM, Rome ME, Clemons WM., Jr Tail-anchor targeting by a Get3 tetramer: The structure of an archaeal homologue. EMBO J. 2012;31(3):707–719. doi: 10.1038/emboj.2011.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alberts B. 2008. Molecular Biology of the Cell (Garland Science, Taylor & Francis Group, New York), 5th ed., p 652.
- 23.Kiekebusch D, Michie KA, Essen L-O, Löwe J, Thanbichler M. Localized dimerization and nucleoid binding drive gradient formation by the bacterial cell division inhibitor MipZ. Mol Cell. 2012;46(3):245–259. doi: 10.1016/j.molcel.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saraogi I, Akopian D, Shan S-O. A tale of two GTPases in cotranslational protein targeting. Protein Sci. 2011;20(11):1790–1795. doi: 10.1002/pro.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wereszczynski J, McCammon JA. Nucleotide-dependent mechanism of Get3 as elucidated from free energy calculations. Proc Natl Acad Sci USA. 2012;109(20):7759–7764. doi: 10.1073/pnas.1117441109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 27.Chang Y-W, et al. Interaction surface and topology of Get3-Get4-Get5 protein complex, involved in targeting tail-anchored proteins to endoplasmic reticulum. J Biol Chem. 2012;287(7):4783–4789. doi: 10.1074/jbc.M111.318329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chartron JW, VanderVelde DG, Rao M, Clemons WM., Jr Get5 carboxyl-terminal domain is a novel dimerization motif that tethers an extended Get4/Get5 complex. J Biol Chem. 2012;287(11):8310–8317. doi: 10.1074/jbc.M111.333252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317(1):41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 30.Shan S-O, Schmid SL, Zhang X. Signal recognition particle (SRP) and SRP receptor: A new paradigm for multistate regulatory GTPases. Biochemistry. 2009;48(29):6696–6704. doi: 10.1021/bi9006989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Schaffitzel C, Ban N, Shan S-O. Multiple conformational switches in a GTPase complex control co-translational protein targeting. Proc Natl Acad Sci USA. 2009;106(6):1754–1759. doi: 10.1073/pnas.0808573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akopian D, Dalal K, Shen K, Duong F, Shan S-O. SecYEG activates GTPases to drive the completion of cotranslational protein targeting. J Cell Biol. 2013;200(4):397–405. doi: 10.1083/jcb.201208045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gasper R, Meyer S, Gotthardt K, Sirajuddin M, Wittinghofer A. It takes two to tango: Regulation of G proteins by dimerization. Nat Rev Mol Cell Biol. 2009;10(6):423–429. doi: 10.1038/nrm2689. [DOI] [PubMed] [Google Scholar]
- 34.Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature. 2010;465(7297):435–440. doi: 10.1038/nature09032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peluso P, Shan S-O, Nock S, Herschlag D, Walter P. Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry. 2001;40(50):15224–15233. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- 36.Wu C, Amrani N, Jacobson A, Sachs MS. The use of fungal in vitro systems for studying translational regulation. Methods Enzymol. 2007;429:203–225. doi: 10.1016/S0076-6879(07)29010-X. [DOI] [PubMed] [Google Scholar]
- 37.Rothblatt JA, Meyer DI. Secretion in yeast: Reconstitution of the translocation and glycosylation of alpha-factor and invertase in a homologous cell-free system. Cell. 1986;44(4):619–628. doi: 10.1016/0092-8674(86)90271-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.