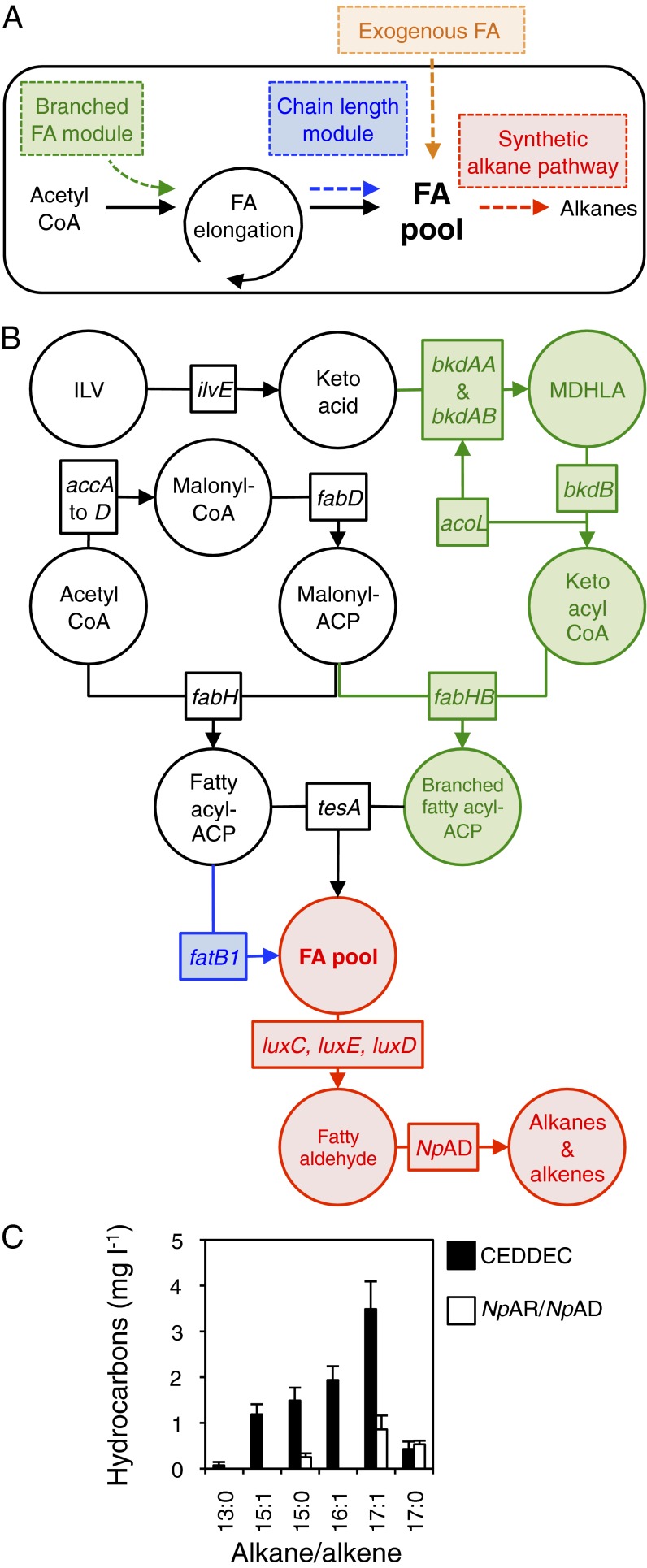
Fig. 1.
Production of fuel-grade alkanes from FAs by engineered E. coli. (A) Overview: three independent modifications to the FA pool (two genetic and one by exogenous supplementation) resulted in predictable changes to the output of cells engineered to produce alkane fuels via the artificial pathway. (B) Details of the genetic modifications implemented: the synthetic alkane biosynthetic pathway (red) was engineered through the coexpression of the reductase (luxC), synthetase (luxE), and transferase (luxD) genes coding for the FAR complex from the P. luminescens luciferase operon and the gene encoding NpAD. For construct details, see Fig. S1. Branched-chain FAs (green) were produced by expression of genes coding for the BCKD complex (bkdAA, bkdAB, bkdB, and acoL, which code for E1α, E1β, E2, and E3 subunits, respectively) and fabHB encoding KASIII. Isoleucine, leucine, and valine (ILV) can be converted to α-keto acids through endogenous branched-chain amino acid aminotransferase activity (encoded by ilvE). MDHLA represents either S-(3-methylbutanoyl)-dihydrolipoamide E, S-(2-methylpropanoyl)-dihdrolipoamide E, or S-(2-methylbutanoyl)-dihydrolipoamide E, the products of isoleucine, leucine, or valine breakdown, respectively (Fig. S2). The E1α, E1β, and E2 subunits convert keto acids to keto acyl-CoA, whereas the E3 subunit recycles the lipoamide-E cofactor. All branched-chain components were from B. subtilis. FA chain length was altered through expression of the FatB1 thioesterase from C. camphora (blue). (C) Hydrocarbons produced by cells expressing the synthetic alkane pathway (CEDDEC) or the cyanobacterial alkane pathway (AR and AD from N. punctiforme) without modifications to the fatty acid pool. n = 6 biological replicates; error bars represent SE of mean. Cells were grown and induced as detailed in Experimental Procedures.