Abstract
Pseudohypoaldosteronism type II (PHAII) is a rare Mendelian syndrome featuring hypertension and hyperkalemia resulting from constitutive renal salt reabsorption and impaired K+ secretion. Recently, mutations in Kelch-like 3 (KLHL3) and Cullin 3 (CUL3), components of an E3 ubiquitin ligase complex, were found to cause PHAII, suggesting that loss of this complex’s ability to target specific substrates for ubiquitination leads to PHAII. By MS and coimmunoprecipitation, we show that KLHL3 normally binds to WNK1 and WNK4, members of WNK (with no lysine) kinase family that have previously been found mutated in PHAII. We show that this binding leads to ubiquitination, including polyubiquitination, of at least 15 specific sites in WNK4, resulting in reduced WNK4 levels. Dominant disease-causing mutations in KLHL3 and WNK4 both impair WNK4 binding, ubiquitination, and degradation. WNK4 normally induces clearance of the renal outer medullary K+ channel (ROMK) from the cell surface. We show that WT but not mutant KLHL3 inhibits WNK4-induced reduction of ROMK level. We show that PHAII-causing mutations in WNK4 lead to a marked increase in WNK4 protein levels in the kidney in vivo. These findings demonstrate that CUL3–RING (really interesting new gene) ligases that contain KLHL3 target ubiquitination of WNK4 and thereby regulate WNK4 levels, which in turn regulate levels of ROMK. These findings reveal a specific role of CUL3 and KLHL3 in electrolyte homeostasis and provide a molecular explanation for the effects of disease-causing mutations in both KLHL3 and WNK4.
Keywords: proteomics, Gordon syndrome, Kir1.1
Hypertension affects 1 billion people worldwide and is a principal reversible risk factor for cardiovascular disease. Identification of the causes of rare Mendelian forms of hypertension has demonstrated the key role of increased renal salt reabsorption in the pathogenesis of this common disease (1, 2).
Among Mendelian hypertensive syndromes, pseudohypoaldosteronism type II (PHAII, also known as familial hypertensive hyperkalemia, Gordon syndrome, OMIM no. 145260) is particularly interesting because it has revealed previously unrecognized physiology involved in orchestrating the activities of different electrolyte flux pathways (3). The kidney is exposed to elevated levels of the steroid hormone aldosterone in two distinct physiologic conditions. Intravascular volume depletion activates the renin-angiotensin system, leading to increased angiotensin II (AII) levels. AII binds to its receptor in adrenal glomerulosa, leading to aldosterone secretion. In this setting, aldosterone signaling leads to a marked increase in renal Na-Cl reabsorption, defending intravascular volume. In the setting of hyperkalemia, high plasma K+ levels depolarize glomerulosa cells, directly producing aldosterone secretion. In this case, aldosterone signaling supports increased electrogenic Na+ reabsorption, providing the electrical driving force for K+ secretion, restoring normal plasma K+ levels. The kidney must be able to distinguish between these two conditions to mount the appropriate physiologic response. In PHAII, the kidney cannot make this distinction appropriately and constitutively reabsorbs Na-Cl at the expense of impaired K+ secretion.
Mutations in four genes have been identified to cause PHAII (4, 5). Two encode the serine-threonine kinases WNK1 (with no lysine kinase 1) and WNK4 (4). Disease-causing mutations in WNK4 are missense mutations that cluster in a short, highly acidic domain of the protein, whereas mutations in WNK1 are large deletions of the first intron that increase WNK1 expression. Biochemistry, cell biology, and animal model studies have demonstrated that WNK4 regulates the balance between renal Na-Cl reabsorption and K+ secretion, with missense mutations found in patients with PHAII promoting increased levels of the renal Na-Cl cotransporter NCC and decreased levels of renal outer medullary K+ channel ROMK (Kir1.1; encoded by KCNJ1), a K+ channel required for normal renal K+ secretion (3, 6–11). WNK4 has been shown to lie downstream of AII signaling (12). AII is the only hormone specific for volume depletion, suggesting that WNK4 mutations phenocopy constitutive AII signaling in the kidney. Nonetheless, the biochemical mechanism by which WNK4 missense mutations change its activity has been unknown.
Recently, we identified mutations in two partners in a cullin-RING (really interesting new gene) E3 ubiquitin ligase (CRL) complex, Kelch-like 3 (KLHL3) and Cullin 3 (CUL3), that explain about 80% of families with PHAII (5). CUL3 is a scaffold protein that assembles a complex that targets specific proteins for ubiquitination. This complex includes a RING E3 ubiquitin ligase and one of a family of more than 50 targeting molecules that bind to CUL3 via amino-terminal bric-a-brac tramtrack broad complex (BTB) domains (13). KLHL3 is one of these targeting proteins. In addition to its N-terminal BTB domain, KLHL3 has a C-terminal six-bladed Kelch domain; these β-propeller structures commonly bind to specific target proteins (14). Mutations in KLHL3 in PHAII are either recessive or dominant (5). Recessive mutations are distributed throughout the protein and include many premature termination, frameshift, and splice site mutations, consistent with loss of function. In contrast, dominant KLHL3 mutations are all missense mutations that cluster in the ends of d-a loops connecting the outermost (d) beta-strand of one Kelch propeller blade to the innermost (a) beta-strand of the next blade. These sites all lie on the same surface of the β-propeller and are known to interact with target proteins in related paralogs (14). CUL3 mutations are all heterozygous, predominantly de novo mutations, and all result in skipping of exon 9, leading to an in-frame 57-aa deletion (5).
From the observation that recessive mutations in KLHL3, dominant mutations in KLHL3, and dominant mutations in CUL3 all result in phenocopies of the same disease, we inferred that all of these mutations likely produce loss of ubiquitination of specific proteins normally targeted by KLHL3. The identity of these proteins was unknown. We report herein that WNK4 is a direct target for ubiquitination by CUL3–KLHL3 CRL complexes and that this ubiquitination leads to degradation and reduced levels of WNK4. We show that disease-causing dominant mutations in either KLHL3 or WNK4 inhibit binding, ubiquitination, and degradation of WNK4, resulting in higher WNK4 levels in mammalian cells and in vivo. This increase in WNK4 level is sufficient to increase inhibition of one of WNK4’s known targets, ROMK. These findings demonstrate that CUL3–KLHL3 and WNK4 act together in the same biochemical pathway and provide a molecular explanation for the mechanism of dominant KLHL3 and WNK4 mutations.
Results
KLHL3 Binds to CUL3, WNK1, and WNK4.
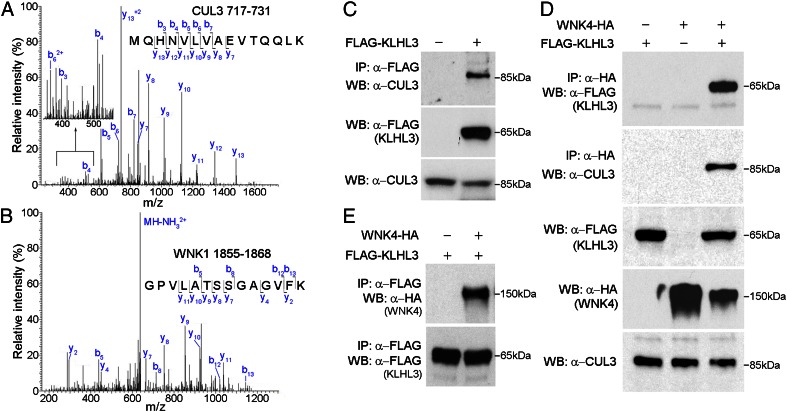
To gain insight into the binding partners of KLHL3, we expressed full-length KLHL3 with a FLAG epitope tag at the amino terminus (FLAG-KLHL3) in COS-7 (monkey kidney-derived) cells, prepared cell lysates, and immunoprecipitated with anti-FLAG antibodies. The products were digested with trypsin and analyzed by liquid chromatography and tandem MS (LC-MS/MS). Peptides were assigned from the precise match of predicted and observed m/z ratios of precursor ions and their product fragment ions. The results identified only three proteins that were found in each of three independent experiments. Not surprisingly, in addition to KLHL3, we found its anticipated CRL partner CUL3 (Fig. 1A and Table S1). In addition, however, we also identified WNK1 (Fig. 1B and Table S2). Mascot protein scores were 793 and 246 for CUL3 and WNK1, respectively. This interaction of CUL3 with KLHL3 was also shown by Western blotting of the products of KLHL3 immunoprecipitation (IP) with anti-CUL3 antibodies (Fig. 1C).
Fig. 1.
Association of WNK kinases with CUL3–KLHL3 complex. (A and B) KLHL3 binds to endogenous CUL3 and WNK1. Products of IP of FLAG-KLHL3 from COS-7 cell lysates were analyzed by LC-MS/MS. Representative MS/MS spectra from peptides mapping to CUL3 (Macaca mulatta, amino acids 717–731) (A) or WNK1 (Macaca mulatta, amino acids 1855–1868) (B) are shown (all peptides in Tables S1 and S2). (C) IP of cell lysates with anti-FLAG (KLHL3) was followed by Western blotting (WB). CUL3 is pulled down only when FLAG-KLHL3 is expressed. (D) Cell lysates expressing indicated proteins were immunoprecipitated with anti-HA, followed by Western blotting. When WNK4-HA is coexpressed with KLHL3, both KLHL3 and CUL3 are immunoprecipitated, whereas neither CUL3 nor KLHL3 is detected in IP products in the absence of KLHL3. (E) Cell lysates expressing the indicated proteins were immunoprecipitated using FLAG antibody to IP FLAG-KLHL3, followed by Western analysis with antibody to HA to detect WNK4-HA (Upper) or FLAG to detect KLHL3 (Lower). IP of FLAG-KLHL3 pulls down WNK4.
We next tested whether WNK4, which is not endogenously expressed in COS-7 cells, might also interact with this ubiquitin ligase complex by expressing full-length WNK4 tagged with the HA epitope at the C terminus (WNK4-HA) in COS-7 cells with or without FLAG-KLHL3. In the presence of FLAG-KLHL3, IP of WNK4-HA with anti-HA pulled down both FLAG-KLHL3 and CUL3 (Fig. 1D). The reciprocal experiment demonstrated that IP of FLAG-KLHL3 with anti-FLAG pulled down CUL3 and WNK4 (Fig. 1E). The ability of anti-HA (WNK4) to pull down CUL3 was dependent upon coexpression of KLHL3, consistent with a physical interaction between KLHL3 and WNK4. Collectively, these results demonstrate that WNK4 can be found in a complex with KLHL3 and CUL3, consistent with the idea that a CUL3–KLHL3 complex might target WNKs for binding and ubiquitination.
PHAII Mutations in KLHL3 and WNK4 Inhibit Their Interaction.
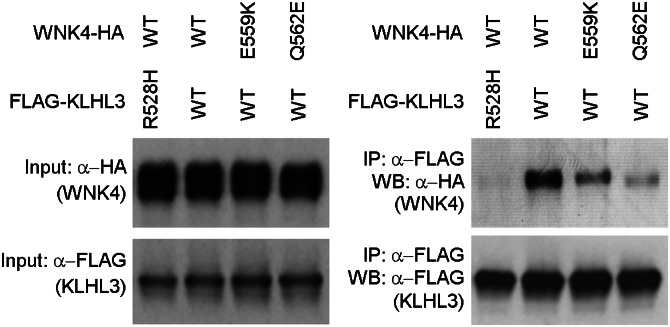
Our findings raise the question of whether dominant PHAII-causing mutations in KLHL3 and WNK4 might prevent binding of WNK4 to KLHL3. Accordingly, we separately expressed constructs bearing WT FLAG-KLHL3 and FLAG-KLHL3 with an R528H missense mutation, which has been found recurrently in unrelated patients with dominant PHAII (5). In parallel, we expressed WT WNK4-HA. We then incubated WT or mutant FLAG-KLHL3 with WNK4-HA and performed IP with anti-FLAG, followed by Western blotting of the resultant protein with anti-HA to detect WNK4. The results demonstrated robust binding of WT KLHL3 to WNK4, but virtually complete loss of binding of KLHL3R528H to WNK4 (Fig. 2). We next performed the analogous experiment using WT FLAG-KLHL3 and either of two dominant WNK4 mutations, E559K or Q562E. The results demonstrate that both of these mutations also impair binding of WNK4 and KLHL3, although to a lesser extent (Fig. 2).
Fig. 2.
PHAII mutations in WNK4 or KLHL3 abrogate WNK4 binding to KLHL3. Cell lysates from cells expressing FLAG-KLHL3WT or FLAG-KLHL3R528H were incubated with lysates from cells expressing WNK4WT-HA, WNK4E559K-HA, or WNK4Q562E-HA followed by anti-FLAG (KLHL3) immunoprecipitation and Western blotting (WB). PHAII mutations in KLHL3 and WNK4 eliminate or reduce binding of WNK4 to KLHL3.
We similarly performed MS on protein immunoprecipitated from COS-7 cells expressing KLHL3R528H. In triplicate experiments, we detected CUL3 in the complex as before, but did not detect WNK1 in any experiment, consistent with the KLHL3 mutation leading to loss of WNK1 binding.
The finding that KLHL3 binds WNK kinases and that reciprocal PHAII-causing mutations impair this interaction implicate this interaction in normal physiology and its loss in disease pathogenesis.
KLHL3 Targets WNK4 for Ubiquitination.
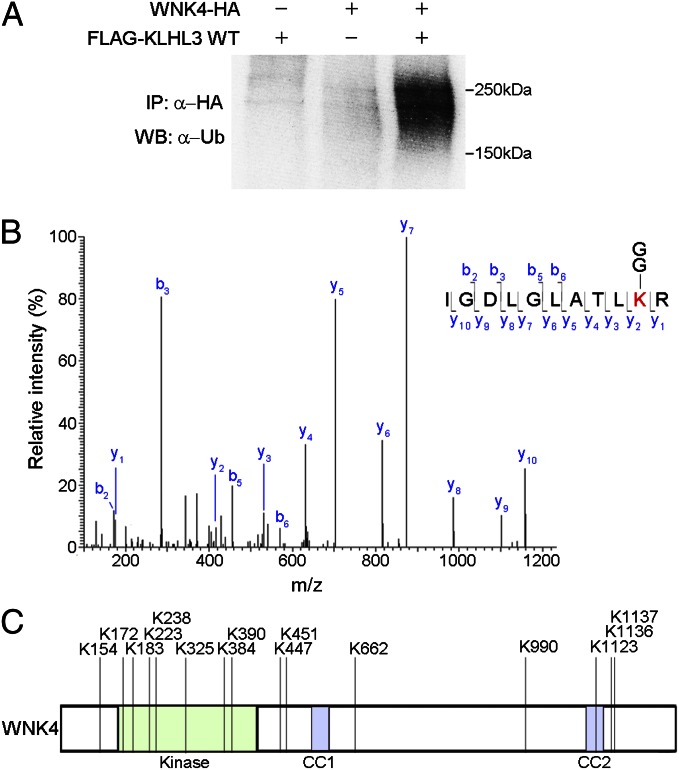
CRLs conjugate a single ubiquitin (monoubiquitination) or polyubiquitin chains (polyubiquitination) to substrates, regulating diverse cellular processes including modulation of protein activity and degradation through the proteasome (15). We evaluated the functional significance of the association between WNK4 and CUL3–KLHL3 by an in vivo ubiquitination assay. WNK4-HA was purified by IP from COS-7 cells with and without coexpression of KLHL3; Western blotting was performed with a monoclonal antibody to ubiquitin. The results demonstrated a robust ladder of ubiquitinated WNK4 when KLHL3 was coexpressed, but not in the absence of KLHL3 (Fig. 3A). These results indicate KLHL3-dependent ubiquitination of WNK4.
Fig. 3.
Identification of ubiquitin conjugation sites in WNK4 by MS. (A) Cell lysates expressing the indicated proteins were immunoprecipitated in denaturing condition with anti-HA, followed by Western blotting (WB) with anti-ubiquitin antibodies. WNK4 is ubiquitinated in the presence but not the absence of KLHL3. (B) MS/MS spectrum of purified ubiquitinated WNK4 showing assignment of the peptide containing ubiquitinated K325 in the WNK4 kinase domain. The precursor ion, 635.882+, was selected and produced the fragment ion spectrum shown. Specific y and b fragment ions allowed the identification of K325 with di-glycine modification (shown as GG) that is derived from the C terminus of ubiquitin. (C) Ubiquitination sites of WNK4 in COS-7 cells expressing KLHL3. Sites are numbered according to their position in mouse sequence. The domains of WNK4 are indicated and lysines that are ubiquitinated are shown. Peptides and Mascot scores are shown in Table 1. CC, coiled-coil domain.
We used LC-MS/MS analysis to map ubiquitination sites in WNK4. Trypsin digestion of the ubiquitinated proteins produces signature peptides containing lysine with a di-glycine remnant that is derived from the carboxyl terminus of ubiquitin covalently attached to the target (16). We prepared lysates from COS-7 cells expressing HA-tagged ubiquitin, FLAG-KLHL3, and Myc-tagged WNK4 (WNK4-Myc). The lysates were immunoprecipitated using anti-HA antibody to pull down ubiquitinated proteins (Fig. S1); WNK4 was then purified by fractionation via SDS/PAGE. This purified WNK4 was then analyzed by MS. This procedure enabled the identification of 15 ubiquitination sites in WNK4 (Fig. 3 B and C and Table 1). These are distributed throughout the protein, including seven in the kinase domain and one in the second coiled-coil domain. Among the analyzed peptides, we also identified the signature of Lys-48–linked polyubiquitin (Mascot ion score 84; ion mass 1459.77) (Fig. S2). This constitutes evidence of polyubiquitination. The same experiment performed in lysates of cells not expressing KLHL3 provided no evidence of WNK4 ubiquitination. These results confirm that WNK4 is directly ubiquitinated in the presence of KLHL3 and identify ubiquitination at many sites.
Table 1.
Ubiquitinated peptides in WNK4
| Ion score | Peptide sequence | Ubiquitination | Observed mass (Da) | Calculated mass (Da) |
| 109 | GVHVELAEEDDGEK*PGLK | K447 | 2,034.98 | 2,034.99 |
| 93 | IGDLGLATLK*R | K325 | 1,269.74 | 1,269.74 |
| 88 | LAPISEEGK*PQLVGR | K990 | 1,706.93 | 1,706.93 |
| 78 | QK*HLSEVEALQTLQK | K1123 | 1,865.00 | 1,865.00 |
| 77 | VTSGTK*PNSFYK | K390 | 1,441.72 | 1,441.72 |
| 67 | FYDSWK*SVLR | K238 | 1,413.70 | 1,413.70 |
| 53 | K*VTSGTK*PNSFYK | K384, K390 | 1,683.86 | 1,683.86 |
| 52 | K*EIEDLYSR | K1137 | 1,265.62 | 1,265.62 |
| 51 | HLSEVEALQTLQK*K | K1136 | 1,736.94 | 1,736.94 |
| 51 | GVHVELAEEDDGEK*PGLK*LWLR | K447, K451 | 2,717.37 | 2,717.38 |
| 49 | REQEEK*EDTETQAVATSPDGR | K154 | 2,489.13 | 2,489.13 |
| 43 | NPVK*TLR | K662 | 940.54 | 940.54 |
| 38 | GSFK*TVYR | K183 | 1,070.55 | 1,070.55 |
| 37 | FSEEVEMLK*GLQHPNIVR | K223 | 2,239.14 | 2,239.14 |
| 33 | YLK*FDIEIGR | K172 | 1,366.72 | 1,366.72 |
K, ubiquitinated lysine.
Polyubiquitination and Down-Regulation of WNK4 Are Abrogated by R528H Substitution in KLHL3.
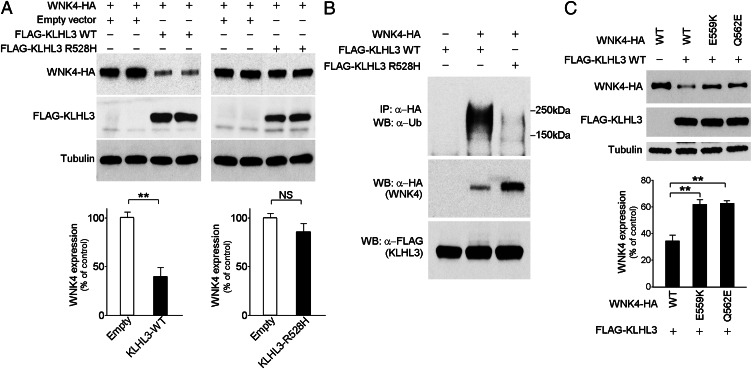
The ability of KLHL3 to promote WNK4 polyubiquitination strongly suggests that this modification should promote WNK4 degradation by the 26S proteasome, which should lead to reduction of WNK4 levels in COS-7 cells. Accordingly, we quantitated WNK4 expression levels in the absence or presence of KLHL3. As shown in Fig. 4A, WNK4 levels were reduced by 60% by coexpression of WT KLHL3 (P = 0.0009).
Fig. 4.
KLHL3WT, but not KLHL3R528H reduces WNK4 levels. (A) Western blots of cell lysates show markedly reduced levels of WNK4 when KLHL3WT but not KLHL3R528H is coexpressed. An empty vector was used as control to equalize the total amount of DNA in transfections lacking KLHL3. (Lower) Bar graphs show quantitation of the results of four independent experiments in each condition. **P < 0.01. (B) COS-7 cell lysates expressing the indicated proteins were immunoprecipitated with anti-HA, followed by Western blotting (WB) with anti-ubiquitin antibodies. Polyubiquitination of WNK4 was markedly reduced in the presence of KLHL3R528H. In addition, WNK4 level was reduced in cells expressing KLHL3WT compared with KLHL3R528H. Representative results of triplicate experiments are shown. (C) Western blots of lysates of cells expressing FLAG-KLHL3WT and either WNK4WT-HA, WNK4E559K-HA, or WNK4Q562E-HA are shown. Mutant WNK4s show significantly highly levels compared with WT. Bar graph shows the results of quantitation (n = 4). Data are expressed as mean ± SEM.
From the binding studies shown, we expect that PHAII mutations that reduce binding of KLHL3 and WNK4 should abrogate this down-regulation of WNK4. Coexpression of WNK4 with KLHL3R528H showed no significant reduction in WNK4 levels compared with the absence of KLHL3 (P = 0.13; Fig. 4A). Furthermore, the polyubiquitination of WNK4 detected in the presence of WT KLHL3 was virtually abolished by the R528H mutation in KLHL3 (Fig. 4B, Top). Similarly, expression of KLHL3WT with PHAII-mutant forms of WNK4 (WNK4E559K and WNK4Q562E) resulted in loss of WNK4 ubiquitination and higher WNK4 levels (Fig. 4C and Fig. S3). These findings indicate that PHAII-causing mutations in both KLHL3 and WNK4 cause loss of the ability of KLHL3 to induce ubiquitination and degradation of WNK4.
KLHL3 Increases Membrane ROMK Expression Through WNK4 Inhibition.
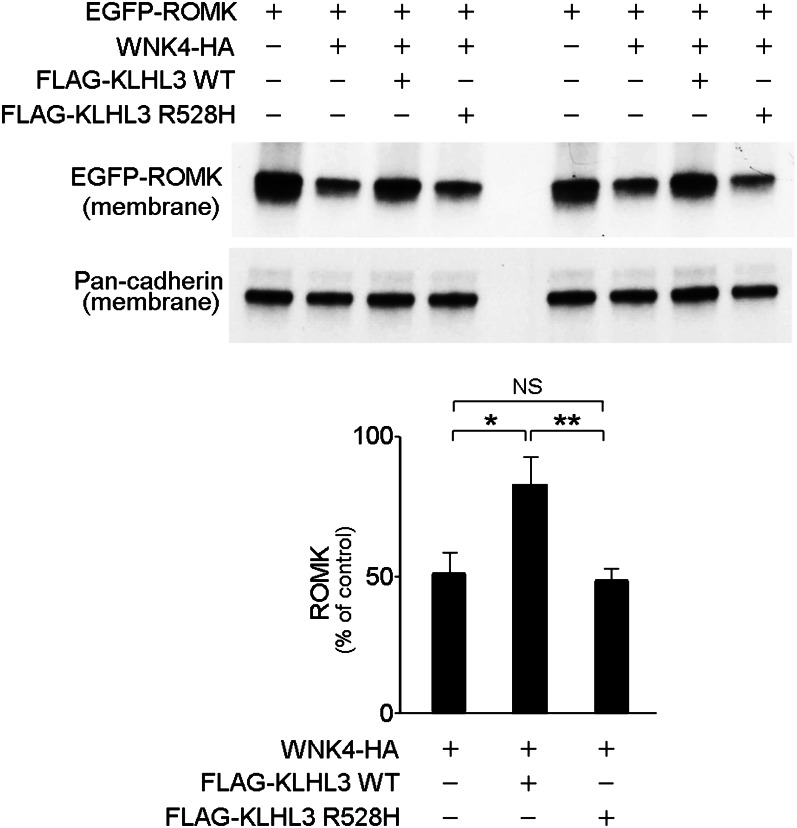
We next evaluated the downstream physiological effect of the interaction between WNK4 and KLHL3. WNK4 inhibits the K+ channel ROMK by reducing the number of channels at the cell surface (6), and PHAII-mutant WNK4 shows increased inhibition of ROMK. To test whether KLHL3 modulates this effect, we expressed ROMK tagged with EGFP in COS-7 cells and quantitated its membrane expression in the presence or absence of WNK4 and KLHL3. As expected, WT WNK4 produced a marked reduction in the expression of membrane EGFP-ROMK (Fig. 5). However, coexpression of WT KLHL3 abrogated the effect of WNK4 and caused a 60% increase in EGFP-ROMK (P < 0.05 vs. WNK4 alone; Fig. 5). In contrast, coexpression of KLHL3R528H failed to reverse WNK4’s inhibition of EGFP-ROMK. These results show that KLHL3 increases ROMK expression by reducing levels of WNK4.
Fig. 5.
KLHL3 abrogates WNK4’s inhibition of ROMK expression. The indicated proteins were expressed, and levels of EGFP-ROMK in the membrane fraction were analyzed by Western blotting (Upper). Cadherin was used as a loading control (Lower). KLHL3WT inhibits WNK4-dependent reduction of ROMK level, and effect lost with KLHL3R528H. Results from biological replicates are shown and bar graphs show the results of quantitation (n = 4). Data are expressed as means ± SEM; *P < 0.05, **P < 0.01.
PHAII Mutation Increases WNK4 Expression in Vivo.
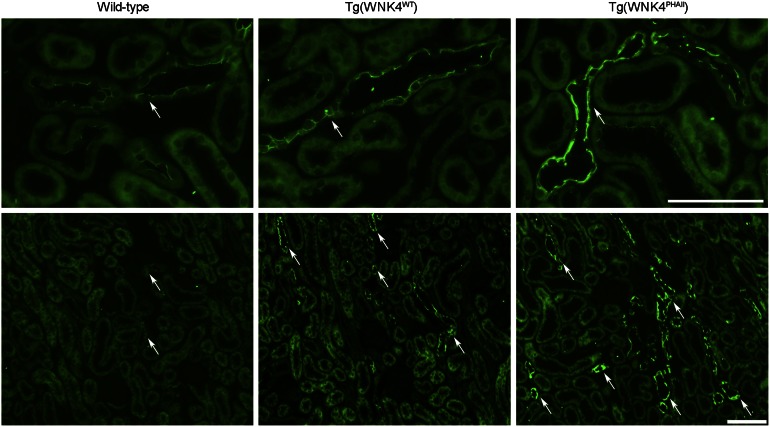
These findings suggest that at least part of the mechanism of PHAII mutations in WNK4 is that WNK4 escapes degradation, resulting in higher levels. We tested this possibility by comparing expression of WNK4 in WT mice and mice carrying a single copy of a bacterial artificial chromosome harboring the WNK4 genomic locus encoding either WNK4WT or WNK4Q562E (11). The latter mice develop hypertension and hyperkalemia like humans who carry this mutation (11). We analyzed WNK4 expression levels by immunostaining of kidney with specific WNK4 antibodies (4). Consistent with prior studies, WNK4 was detected predominantly in distal convoluted tubule (DCT), collecting duct (CD), and connecting tubule (CNT) in the kidney. We found a dramatic and consistent increase in WNK4 staining intensity in mice harboring WNK4Q562E compared with WT littermates or mice harboring the WT WNK4 transgene (Fig. 6). These data are consistent with the Q562E mutation preventing KLHL3-directed degradation of WNK4 and with this effect playing a role in the pathogenesis of PHAII.
Fig. 6.
Renal expression of WNK4 in WT, Tg(WNK4WT), and Tg(WNK4PHAII) mice. Kidneys from 3- to 5-mo-old mice of indicated genotypes (n = 3 of each) were fixed by in vivo perfusion and sectioned and stained with anti-WNK4. (Upper) High-power and (Lower) low-power images. Intensity of WNK4 staining (arrows) was consistently increased in Tg(WNK4PHAII) mice compared with WT littermates or Tg(WNK4WT). (Scale bars, 100 μm.)
Discussion
These results provide insight into the molecular mechanisms by which the expression and function of WNK4, a major determinant of the balance between renal salt reabsorption and K+ secretion, is regulated. CUL3–KLHL3 ubiquitin ligases bind and target WNK4 for ubiquitination and degradation. These findings provide a biochemical link between CUL3, KLHL3, and WNK4, explaining the phenotypic similarity resulting from mutations in all these genes. The functional importance of this interaction is clear because PHAII mutations in either KLHL3 or WNK4 impair this interaction, reduce WNK4 ubiquitination, and result in increased WNK4 levels and increased inhibition of ROMK. This finding is shown to extend to a mouse model of PHAII which shows a marked increase in WNK4 in mice bearing an extra copy of PHAII-mutant, but not WT WNK4.
The sites of PHAII mutations in the Kelch domain of KLHL3 and the acidic domain of WNK4 likely identify specific sites in KLHL3 and WNK4 that are required for these interactions. This inference is supported by data from the crystal structure of another Kelch-domain targeting molecule, Kelch-like ECH-associated protein 1 (KEAP1), and one of its targets, Nuclear factor erythroid 2-related factor 2 (NRF2). This interaction occurs between basic amino acids in d-a loops of KEAP1 and an acidic domain in NRF2 (14), highly similar to the interactions between basic residues in the d-a loops of KLHL3 and an acidic domain of WNK4 indicated by the genetic and biochemical data herein. The dependence of CUL3–KLHL3-mediated degradation of WNK4 on the WNK4 acidic domain provides an explanation for the effects of mutations in this domain on WNK4 function.
Because WNK1 has an acidic domain nearly identical to WNK4, it is not surprising that WNK1 also interacts with KLHL3, and that this binding is also lost in the presence of the KLHL3R528H mutation. The functional consequences of this binding have not been explored.
These results suggest potential mechanisms for the observed genotype–phenotype correlation in which patients with KLHL3 mutations have more severe disease than those with WNK4 mutations (5). With dominant WNK4 mutations, only one WNK4 allele is expected to be protected from degradation; with recessive or dominant KLHL3 mutations, both WNK4 alleles would be protected from degradation and WNK1 levels might also be increased, potentially explaining the greater severity of the KLHL3 mutations.
During preparation of this article, Ohta et al. reported related findings demonstrating physical interactions of WNK1, WNK4, and KLHL3 (17). Interestingly, this study did not find altered expression of WNK1 resulting from this interaction. Ohta et al. did not report effects of KLHL3 on ubiquitination or expression of WNK4 or the impact on downstream targets of WNK4 in mammalian cells or in vivo. The results of our study and Ohta et al. (17) are generally complementary.
Given the evidence that WNK4 lies downstream of AII signaling and that PHAII mutations in WNK4 phenocopy effects of this signaling pathway (12), a major question has been how AII signaling normally modifies WNK4 activity. The discovery of this link between WNK kinases and CUL3–KLHL3 suggests that AII signaling likely regulates WNK kinase levels by regulating ubiquitination by CUL3–KLHL3. This could be achieved by modifying CUL3–KLHL3, WNK4, or both. Further work will be required to explore these possibilities.
Materials and Methods
Clones and Mutagenesis.
The expression plasmid encoding human KLHL3 was obtained from Origene. A FLAG epitope tag was introduced in-frame by PCR. The R528H mutation was introduced into KLHL3 using the QuikChange site-directed mutagenesis system (Stratagene). WNK4 and ROMK constructs were as described previously (6, 7).
LC-MS/MS.
MS was performed at the Yale W. M. Keck Foundation Biotechnology Resource Laboratory as described previously (18). HA-tagged WNK4 expressed in COS-7 cells was immunoprecipitated using anti-HA antibodies, the product was digested with trypsin, and peptide mixtures were fractionated by HPLC interfacing an electrospray ionisation quadrupole time-of-flight mass spectrometer. All MS/MS spectra were searched using the Mascot algorithm (19). A protein is considered identified when there are two or more peptide matches to a protein, each with Mascot ion score > 30. The database searched was National Center for Biotechnology Information, nr (non-redundant) database. Searches allowed variable methionine oxidation and carbamidomethylated cysteine, a peptide tolerance of ± 20 ppm and MS/MS fragment tolerance of ± 0.6 Da. For ubiquitination site mapping, di-glycine modification of lysine was also allowed as a variable modification. Details on LC-MS/MS analysis are provided in SI Materials and Methods.
Transient Transfection and IP.
COS-7 cells (American Type Culture Collection) were cultured in DMEM (GIBCO) supplemented with 10% (vol/vol) heat-inactivated FBS. Transient transfection was performed using cationic liposome (Lipofectamine 2000, Life Technologies) followed by incubation for 48 h; cells were then washed in cold PBS and lysed at 4 °C in lysis buffer (10 mM Tris⋅HCl, pH 7.8/150 mM NaCl/1 mM EDTA/1% Nonidet P-40) containing protease inhibitor (Roche). Protein concentrations were equalized by quantitation and incubated with mouse monoclonal anti-FLAG or rabbit monoclonal anti-HA agarose conjugate at 4 °C. Immunoprecipitates were washed and bound protein was eluted by boiling in Laemmli sample buffer. For ubiquitination assay and analysis of ubiquitin conjugation sites, cells were lysed in 1% SDS, 150 mM Na-Cl, and 10 mM Tris⋅HCl, pH 8.0, and immediately boiled. Samples were diluted by adding 4× volume of solution containing 1% Triton-X, 150 mM Na-Cl, 10 mM Tris⋅HCl (pH 8.0), 1 mM EDTA, and 0.5% N-ethylmaleimide. The resultant lysates were immunoprecipitated with anti-HA agarose conjugate as described above.
Western Blotting.
Protein extraction and Western blotting were performed as described previously (20). Membrane/cytoplasmic protein fractionation was performed using reagents purchased from Thermo Scientific (MEM-PER PLUS). The purification of membrane proteins in the membrane fraction was confirmed by the enrichment of cadherin. Lysates and immunoprecipitated proteins were separated on 7.5% (wt/vol) polyacrylamide gel, transferred to nitrocellulose membrane, and immunoblotted with primary antibodies including rabbit anti-HA (Sigma, 1:2,000), anti-FLAG (Sigma, 1:5,000), anti-CUL3 (Abcam, 1:20,000), anti-ubiquitin (Cell Signaling, 1:1,000), anti-GFP (Invitrogen, 1:200), anti-pan-cadherin (Sigma, 1:2,000), or anti-tubulin (Sigma, 1:2,000) antibody. After incubation with peroxidase-conjugated secondary antibody, signals were visualized by chemiluminescence.
Animal Studies and Immunostaining.
Mice were maintained following a protocol approved by the Yale Institutional Animal Care and Use Committee (Protocol 2008-10018). They were fed ad libitum and housed under a 12-h light cycle. Mice transgenic for genomic segments harboring WT (TgWNK4WT) or PHAII mutant WNK4 (Q562E; TgWNK4PHAII) (11) in the C57BL/6 background and WT littermates were studied. Cryosections from perfusion-fixed mouse kidneys were stained with WNK4 antibody (4) (1:100) overnight, and subsequently with affinity-purified secondary antibody conjugated to Alexa Fluor 488 fluorophore.
Statistical Analysis.
The data are summarized as means ± SEM. Unpaired t test was used for comparisons between two groups. For multiple comparisons, statistical analysis was performed by ANOVA with Tukey post hoc tests. P values < 0.05 were considered to be significant.
Supplementary Material
Acknowledgments
We thank Jesse Rinehart, Lynn Boyden, and Carol Nelson-Williams for advice and helpful discussions. This work was supported in part by National Institutes of Health Grants P01-DK017433, P30-DK079310, UL1-RR024139, and P30-DA018343; the Leducq Transatlantic Network on Hypertension; and a fellowship grant from the Uehara Memorial Foundation. R.P.L. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 7535.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304592110/-/DCSupplemental.
References
- 1.Lifton RP, et al. A chimaeric 11 beta-hydroxylase/aldosterone synthase gene causes glucocorticoid-remediable aldosteronism and human hypertension. Nature. 1992;355(6357):262–265. doi: 10.1038/355262a0. [DOI] [PubMed] [Google Scholar]
- 2.Choi M, et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science. 2011;331(6018):768–772. doi: 10.1126/science.1198785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kahle KT, Ring AM, Lifton RP. Molecular physiology of the WNK kinases. Annu Rev Physiol. 2008;70:329–355. doi: 10.1146/annurev.physiol.70.113006.100651. [DOI] [PubMed] [Google Scholar]
- 4.Wilson FH, et al. Human hypertension caused by mutations in WNK kinases. Science. 2001;293(5532):1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 5.Boyden LM, et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature. 2012;482(7383):98–102. doi: 10.1038/nature10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kahle KT, et al. WNK4 regulates the balance between renal NaCl reabsorption and K+ secretion. Nat Genet. 2003;35(4):372–376. doi: 10.1038/ng1271. [DOI] [PubMed] [Google Scholar]
- 7.Wilson FH, et al. Molecular pathogenesis of inherited hypertension with hyperkalemia: The Na-Cl cotransporter is inhibited by wild-type but not mutant WNK4. Proc Natl Acad Sci USA. 2003;100(2):680–684. doi: 10.1073/pnas.242735399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang CL, Angell J, Mitchell R, Ellison DH. WNK kinases regulate thiazide-sensitive Na-Cl cotransport. J Clin Invest. 2003;111(7):1039–1045. doi: 10.1172/JCI17443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kahle KT, et al. Paracellular Cl- permeability is regulated by WNK4 kinase: Insight into normal physiology and hypertension. Proc Natl Acad Sci USA. 2004;101(41):14877–14882. doi: 10.1073/pnas.0406172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamauchi K, et al. Disease-causing mutant WNK4 increases paracellular chloride permeability and phosphorylates claudins. Proc Natl Acad Sci USA. 2004;101(13):4690–4694. doi: 10.1073/pnas.0306924101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lalioti MD, et al. Wnk4 controls blood pressure and potassium homeostasis via regulation of mass and activity of the distal convoluted tubule. Nat Genet. 2006;38(10):1124–1132. doi: 10.1038/ng1877. [DOI] [PubMed] [Google Scholar]
- 12.San-Cristobal P, et al. Angiotensin II signaling increases activity of the renal Na-Cl cotransporter through a WNK4-SPAK-dependent pathway. Proc Natl Acad Sci USA. 2009;106(11):4384–4389. doi: 10.1073/pnas.0813238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prag S, Adams JC. Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals. BMC Bioinformatics. 2003;4:42. doi: 10.1186/1471-2105-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006;25(15):3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12(4):220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng J, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21(8):921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 17.Ohta A, et al. The CUL3-KLHL3 E3 ligase complex mutated in Gordon’s hypertension syndrome interacts with and ubiquitylates WNK isoforms; Disease-causing mutations in KLHL3 and WNK4 disrupt interaction. Biochem J. 451(1):111–122. doi: 10.1042/BJ20121903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rinehart J, et al. Sites of regulated phosphorylation that control K-Cl cotransporter activity. Cell. 2009;138(3):525–536. doi: 10.1016/j.cell.2009.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirosawa M, Hoshida M, Ishikawa M, Toya T. MASCOT: Multiple alignment system for protein sequences based on three-way dynamic programming. Comput Appl Biosci. 1993;9(2):161–167. doi: 10.1093/bioinformatics/9.2.161. [DOI] [PubMed] [Google Scholar]
- 20.Shibata S, et al. Modification of mineralocorticoid receptor function by Rac1 GTPase: Implication in proteinuric kidney disease. Nat Med. 2008;14(12):1370–1376. doi: 10.1038/nm.1879. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.