Abstract
Many common, important diseases are either caused or exacerbated by hyperactivation (e.g., cancer) or inactivation (e.g., heart failure) of the cell division cycle. A better understanding of the cell cycle is critical for interpreting numerous types of physiological changes in cells. Moreover, new insights into how to control it will facilitate new therapeutics for a variety of diseases and new avenues in regenerative medicine. The progression of cells through the four main phases of their division cycle [G0/G1, S (DNA synthesis), G2, and M (mitosis)] is a highly conserved process orchestrated by several pathways (e.g., transcription, phosphorylation, nuclear import/export, and protein ubiquitination) that coordinate a core cell cycle pathway. This core pathway can also receive inputs that are cell type and cell niche dependent. “Broken cell” methods (e.g., use of labeled nucleotide analogs) to assess for cell cycle activity have revealed important insights regarding the cell cycle but lack the ability to assess living cells in real time (longitudinal studies) and with single-cell resolution. Moreover, such methods often require cell synchronization, which can perturb the pathway under study. Live cell cycle sensors can be used at single-cell resolution in living cells, intact tissue, and whole animals. Use of these more recently available sensors has the potential to reveal physiologically relevant insights regarding the normal and perturbed cell division cycle.
Keywords: cell cycle pathway, live cells, sensors, analysis methods, review
the cell division cycle, referred to here as the cell cycle, is a highly conserved pathway that plays a central role in tissue development and cellular homeostasis. The cell cycle consists of three “gap” phases, G0, G1, and G2, that are interspersed between two other phases, i.e., the S (DNA synthesis) phase, where DNA synthesis results in chromosome duplication, and the M (mitosis) phase, where the process of karyokinesis is often coupled to cytokinesis to complete cell division (Fig. 1). Cellular dormancy is when normal cells exit the cell cycle and enter G0. Cells in G0 often display a decrease in overall metabolic activity, reduced rRNA synthesis, decreased translation, and decreased cell size, rendering them resistant to a variety of stresses (2, 116). However, distinguishing between cells in G0 and cells that are arrested in G1 can be technically challenging, and thus many adult nondividing somatic cell types (e.g., cardiomyocytes, neurons) are commonly classified as being in G0/G1 arrest (28, 87, 95, 124).
Fig. 1.
Main phases of the cell cycle.
Many of the core genes that control the cell cycle were first discovered in yeast, and this model organism continues to shed light on new pharmacological agents that perturb this important pathway (72). Dysregulation of the cell cycle can lead to the development and progression of a malignant phenotype. It is thus not surprising that the majority of currently available cancer therapeutics target core aspects of the cell cycle pathway (119).
The cell cycle is controlled by an orchestrated network of genes and their encoded proteins that are temporally regulated by transcription, dynamic protein-protein interactions, and posttranslational modifications (e.g., phosphorylation, ubiquitination). Many mammalian cells require 12–24 h to complete cell division, with differences in cycle time goverened primarily by time spent in the various gap phases (i.e., G0, G1, and G2) (105). This “circadian” nature of cell cycle kinetics is likely influenced by the regulation of core cell cycle genes (e.g., P21, Ccnd1, Ccnb2, Ccna1) by circadian clock genes or their encoded proteins (9). However, in some circumstances, e.g., neural stem cells in vivo, cell cycling times can be much shorter (12 h) or longer (≤15 days or more) depending on cell type and anatomical location (71).
Classic methodology to assess for cell cycle progression, such as quantification of DNA content, DNA synthesis, and immunohistochemical analysis of proliferative marker genes, has enabled many seminal insights into signaling pathways and pharmacological agents that control it. Many of these classic experimental approaches relay on cell fixation and/or synchronization, and thus aspects of cell proliferation cannot be examined by such “broken cell” methods. For example, classic approaches cannot be employed for longitudinal studies examining heterogeneity of cycling kinetics in cultured cells, tissues, or intact animals.
The development of new methods that enable the assessment of cell cycle progression in living cells, tissues, and intact animals has set the stage for unprecedented studies in the area of cell cycle regulation. These tools are revealing new insights into the dynamic temporal and heterogeneous kinetics of the cell cycle. For example, it is now possible to label subpopulations of living cells in each of the main phases of the cell cycle with genetically encoded fluorescent proteins, thereby providing a molecular beacon to pinpoint single cells as they traverse through the cell cycle. This approach enables one to quantify cells in specific phases of the cell cycle over time without perturbing cell or tissue structure, thereby maintaining cell or tissue integrity for downstream applications. For example, using imaging one can simultaneously track and quantify cells in specific phases of the cell cycle while quantifying the duration of each cell in specific phases. Alternatively, these live cell sensors facilitate high-throughput screening of compounds or cDNAs that can induce or inhibit the cell cycle with standard fluorescent plate readers.
Live cell sensors enable in vivo studies that were not previously possible. They can be used to visualize heterogeneity of tumor cell proliferation in vivo as a function of tissue microenvironment and/or drug treatments. These live cell cycle sensors can also be used to isolate and purify cell populations in distinct phases of the cell cycle for genomic studies such as transcriptional profiling or proteomics. Such studies could be used to probe complex gene regulatory networks and/or quantify changes in cellular proteins during specific phases of the cell cycle. Other, more detailed specific examples of the application of these novel reporters are provided and referenced below.
Another application of these novel live cell cycle sensors is in the exciting and burgeoning area of single-cell analysis. The goal of examining cellular heterogeneity over many days or even weeks at the single-cell level was once just a hope. This goal is now attainable with the advent of live cell cycle reporters and rapidly developing technologies, which include single-cell PCR, advances in live cell and whole animal in vivo imaging, and laser capture microdissection. Integration of these approaches has enabled simultaneous multiparameter tracking and quantification of the cell cycle within single cells in vitro and in vivo.
In this review, we first provide a general overview of the core signaling pathway that controls the cell cycle. The fundamental molecular processes at the heart of this pathway, such as DNA synthesis and the expression of proteins during specific phases of the cell cycle, serve as the basis for the development of methods to assess cell cycle progression. We then discuss classic methods that generally rely on broken cell or fixed tissue techniques. Finally, we discuss newer approaches that use cell-permeable or genetically encoded live cell cycle sensors that have enabled studies of the cell proliferation at single-cell resolution in intact cells, tissues, and whole animals.
Several reviews and books on cell cycle biology and techniques have been published (e.g., see Refs. 24, 43, 77, 88, and 119). The purpose of this review is to integrate newer findings and methods in the context of classic ones and their inherent limitations. We anticipate that organizing this review in such a way will provide an intellectual platform for the creative integration of old and new techniques to ask important new questions in this area.
The Core Signaling Pathway that Controls the Cell Cycle
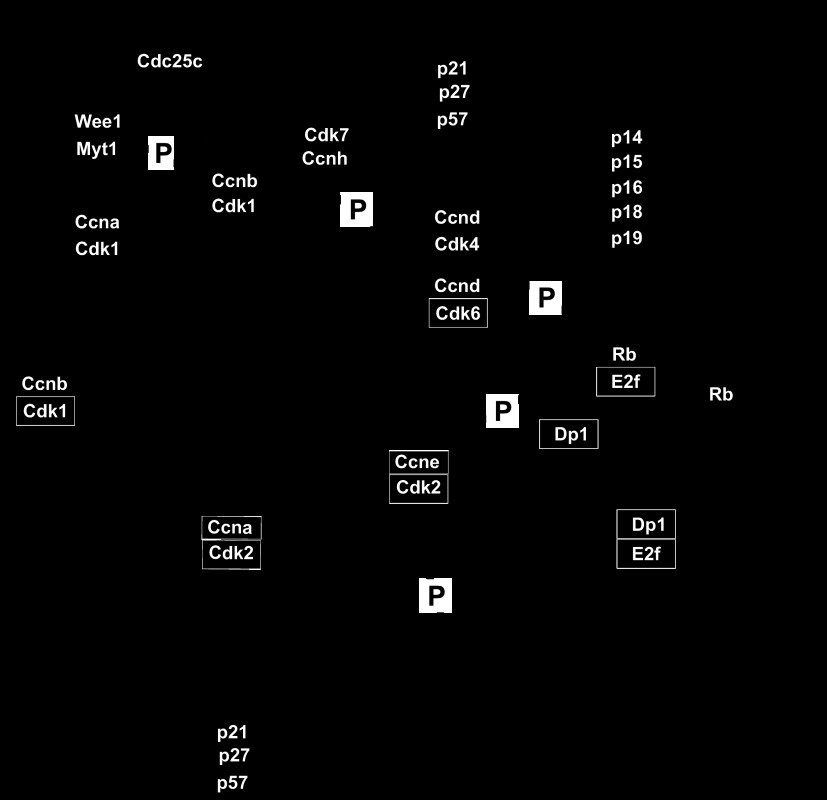
The transition into G1 prepares the cells for DNA synthesis and is controlled by interactions of cyclin-dependent kinase (CDK)4 and CDK6 with their heterodimeric partners: the cyclin D CCND1–3 family members (the choice of partner is poorly understood) (12, 107). The induction of CDK4/6 and CCND proteins facilitates their translocation to the nucleus, where they become further activated by phosphorylation from the CDK-activating kinase (CAK) complex (comprised of CDK1, CDK2, and cyclin A or B). Subsequently, CDK4/CCND1 phosphorylates retinoblastoma (RB) protein family members catalyzing RB exchange with transcription factor Dp1 (Tfdp1) family members for binding to and the subsequent activation of the E2F family of transcription factors. Seven E2F (1–7), two Tfdp (1, 2), and three RB (p-RB, p-107, and p-130) genes have been identified in humans and mice (24, 25). Activated E2F/TFDP heterodimers bind to DNA consensus elements in the promoters of genes that carry out DNA synthesis during the S phase (Fig. 2). CDK2/CCNE heterodimers complete the transition into the S phase by phosphorylation of the initiators of DNA replication among others (74).
Fig. 2.
Simplified pathway representation of the major regulators of cell cycle progression. CDK, cyclin-dependent kinase; CAK, CDK-activating kinase; RB, retinoblastoma; P, phosphorylation; G, gap phase; INK4, inhibitor of cyclin-dependent kinase 4; KIP, kinase inhibitor protein; CIP, cyclin inhibitor protein.
A variety of cell cycle checkpoint genes control progression through the cell division cycle. These CDK inibitors (CKIs) inhibit cell cycle progression by binding to and sequestering/inhibiting CDK activity (18). CKIs are regulated by both transcriptional and posttranscriptional mechanisms (e.g., subcellular sequestration, ubiquitination). They fall into two classes: the INK4 family (p14, p15, p16, p18, and p19) and the KIP/CIP family (p21, p27, and p57). These two families are structurally unrelated and have differing specificities for regulating cell cycle progression (Fig. 2) (11).
P21 and P27(KIP1) negatively regulate CDK activity and also help assemble CCND/CDK4 and CDK6 complexes in G1, thereby both inhibiting and to a smaller extent facilitating cell cycle progression (99). This balance between pro- and antiproliferative signaling is mediated by a complex balance of stoichiometric interactions and changes in cellular compartmentalization and by mechanisms that have yet to be fully elucidated. P21 and P27 promote CCND/CDK4 and CDK6 interactions by stablizing the complexes and by directing the complexes to the nucleus (99). Low concentrations of P21 promote assembly and kinase activity, whereas higher concentrations of P21 inhibit kinase activity (58). The inhibition of CDK4 by P27 depends on the absence or presence of P27 tyrosine phosphorylation, which modifies P27 from a bound inhibitor to a bound noninhibitor (91). During the G1-to-S transition, P27 is ubiquitinated by S phase kinase-associated protein 2 (SKP2) and then targeted for degradation (99). The degradation of P27 is necessary for the transition from G1 into S; this is associated with an increase in E2F-dependent transcriptional activity (64). As discussed below, some of these changes in protein stability and cellular compartmentation have been exploited to generate sensors for assessment of cell cycle regulation.
During the progression through the S phase, CCNA/CDK2 complexes form and phosphorylate several proteins, including cell division cycle 6 protein (CDC6; Fig. 2). CDC6 is involved in the formation of the initiation complex and origin liscensing (40, 46). Mechanisms that prevent DNA rereplication in eukaryotic cells inhibit origin licensing. Origin licensing occurs by the binding of the origin recognition complex, a multisubunit ATPase, to DNA at the replication origins and the recruitment of CDC6 and the chromatin licensing and DNA replication factor 1 (CDT1) protein. These events are required prior to the recruitment and binding of minichromosome maintenance proteins 2–7 onto chromatin (69). The minichromosome maintenance complex, once bound, is the DNA helicase that opens the helix at the replication origin and unwinds the two strands as replication forks travel along the DNA (17).
In G2, the fidelity of chromosome replication is assessed by the DNA damage response pathway as the cell prepares to enter the M phase. A primary function for G2 is to prevent chromosome segregation in the presence of unreplicated or damaged DNA (102). CCNB/CDK1 complexes drive early G2 transition, and their activity is regulated by phosphorylation and ubiquitination (77). If DNA damage is detected, signaling pathways (e.g., activity of ATM/ATR kinases and their downstream substrates CHK1 and CHK2 kinases) lead to the phosphorylation of the phosphatase CDC25C sequestering it in the cytoplasm and preventing it from dephosphorylating and activating nuclear CCNB/CDK1 triggering G2 progression (Fig. 2) (54). Subsequently, the cell enters G2 arrest (91). When DNA replication is complete, CCNA/CDK1 complexes regulate the transistion into the M phase.
During the entry into the M phase, the activity of CDC25C with CDK1 is greater than the activities of opposing kinases WEE1 and MYT1. CDC25C and WEE1 work in positive and negative feedback loops, respectively, to fully activate CCNB/CDK1. The activated CCNA/CDK1 and CCNB/CDK1 complexes phosphorylate substrates that are important for nuclear envelope breakdown and centrosome separation (77). During G2, centrosomes prepare for duplication so that they can organize the spindle poles during mitosis. In mammalian cells, centrosomes are comprised of two centrioles surrounded by pericentriolar material. The single centrosome duplicates prior to cell entry into mitosis. The resulting daughter centrosomes create the poles of the spindle once the nuclear envelope is broken down in mitosis (41).
Mitosis occurs in five phases: prophase, prometaphase, metaphase, anaphase, and telophase. During mitosis the CCNB/CDK1 complex phosphorylates CDC25C, and this creates a positive feedback loop to induce activation of CCNB/CDK1 at the G2/M transition (Fig. 2) (45, 83). The CCNB/CDK1 complex helps initiate various events in mitosis. In prophase, the chromosomes condense and centrosomes promote nuclear envelope breakdown (4). Here, the CCNB/CDK1 complex phosphorylates the centrosome-associated motor protein Eg5 (8), which leads to centrosome separation. In nuclear lamina breakdown, the lamins disperse, the metaphase mitotic spindle is created, and the lamins complete their dispersal before the sister chromatids separate in anaphase (81). The CCNB/CDK1 complex helps with breaking down the nuclear lamina and cell rounding that occurs by the disassembly and reassembly of microfilaments (121). The chromosome condensation that occurs in prophase is controlled in part by activity of mitotic histone H3 kinases. Aurora kinase family members and other kinases can phosphorylate the NH2 terminus of H3 at Ser10 and Ser28 (39) to initiate chromosome condensation. As discussed below, both Aurora kinases and phospho-H3 (Ser10) are commonly used immunohistochemical markers for cell proliferation. CCNA and CCNB must be degraded to exit mitosis; this degradation is accomplished by a ubiquitin-mediated pathway that is regulated by the anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase (31, 76). This cell cycle phase-dependent change in protein stability by APC/C has also been harnessed to generate live cell cycle sensors, as discussed below.
Cytokinesis, the physical separation of a single cell into two daughter cells, is regulated by the Rho proteins, which are part of the Ras family of GTPases. The Rho proteins are involved with positioning the cell division plane as well as the spatial and temporal regulation of the contractile ring (49). The RhoA protein is concentrated at the division plane before cell division occurs and is activated in the equatorial plane. The contractile ring is formed from actin filaments and myosin (82). RhoA promotes actin assembly as well as myosin II motor activation, and then the actin filaments elongate, as is required for the creation of the contractile ring (49).
Classic Methodology to Assess for Cell Cycle Progression
Cell counting and incorporation of labeled nucleoside analogs.
The most direct approach for assessing an end point of cell cycle activity, i.e., cytokinesis, is to simply count cells over time. In light of the fact that some cells become polynucleated and/or polyploid in vitro and in vivo and under stress [e.g., cardiomyocytes (59, 65, 104, 118)], determining total cell number is necessary when assessing cell proliferation by other methods (e.g., increased DNA synthesis, as discussed below). Cell counting is often conducted in the presence of a dye (e.g., trypan blue), which is used to identify cells with compromised cellular membranes. This enables one to exclude apoptotic or necrotic cell populations. These types of studies are usually conducted with a hemocytometer that employs a grid system that bounds a defined volume, thereby enabling cell density determination.
Coulter counters are also able to assess for cell number while simultaneously recording distributions of cell size. These machines work on the principle that cells are nonconducting particles that disrupt a current when passing through a channel. The size of current disruption is proportional to cell size. In recent years, a number of smaller and cheaper instruments that are based on the coulter principle (e.g., Scepter; EMD-Millipore) or image quantification Countess (Invitrogen) have become available as alternatives to coulter counters.
Although counting cell numbers as a function of time is a straightforward method for assessing cell proliferation, in some situations it may not be sensitive enough to detect small but significant changes in proliferation rates. Moreover, it cannot be used to assess cell proliferation in vivo. Cell-permeable nucleoside analogs {e.g., [3H]thymidine, bromodeoxyuridine (BrdU)} are a sensitive means to assess for DNA synthesis as an indication of cell proliferation (63, 79). These analogs are incorporated into newly synthesized DNA during S phase progression. Importantly, they can be used in vitro and in vivo in pulse chase experiments to assess cell proliferation during a defined window of time (79, 104). The use of BrdU has largely replaced that of [3H]thymidine since the latter requires radioactive handling procedures that can pose hurdles in some laboratory settings. Moreover, the use of BrdU facilitates multiparameter assessment (e.g., immunohistochemical staining for cellular antigens as well as changes in DNA synthesis).
Other “BrdU-like” nucleoside analogs with modified halogen moieties such as chlorodeoxyuridine or iododeoxyuridine have been developed to enable sequential S phase cell labeling in vitro and in vivo. This approach can be useful for identifying cells that have undergone two rounds of DNA synthesis (115, 117). These analogs label cells that are traversing S phase with equal efficiency when administered in equimolar concentrations, thereby enabling both qualitative and quantitative studies.
BrdU and possibly other halogenated thymidine analogs have limitations that include adverse effects on cell proliferation and viability (63). In addition, in some cellular settings (e.g., neurons), incorporation of nucleoside analogs occurs in cells undergoing DNA repair, turnover, and/or apoptosis (5, 57). It is postulated that a DNA damage response is triggered in terminally differentiated neurons subjected to death-inducing stimuli in vitro or in vivo as well as in Alzheimer's disease (33). A variety of death-inducing stimuli can upregulate cyclins, CDKs, and DNA synthesis, thereby increasing BrdU incorporation in postmitotic neurons undergoing apoptosis (5, 33). Staining for BrdU, chlorodeoxyuridine, and iododeoxyuridine with anti-sera also requires strong denaturing conditions such as the use of concentrated HCl or mixtures of methanol and acetic acid and high heat, which can degrade the structure of the specimen. These detection conditions can disrupt epitopes in colabeling experiments.
In light of these limitations, 5-ethynyl-2′-deoxyuridine (EdU) was developed as an alternative to BrdU (94). EdU can be detected with fluorescent azides in a Cu(I)-catalyzed “click chemistry” reaction that is highly sensitive and much faster than BrdU detection. EdU does not require an antibody, and thus the label has a higher diffusion rate that penetrates tissue much more effectively. Moreover, denaturation of the specimen is not required, thus maintaining cell epitope integrity for colabeling experiments. However, a variety of fluorophores (e.g., GFP, R-phycoerythrin) are diminished by Cu(I)-catalyzed detection of EdU; alternative EdU detection strategies should be employed when codetection of these molecules is desired (6).
Cell-impermeable DNA stains.
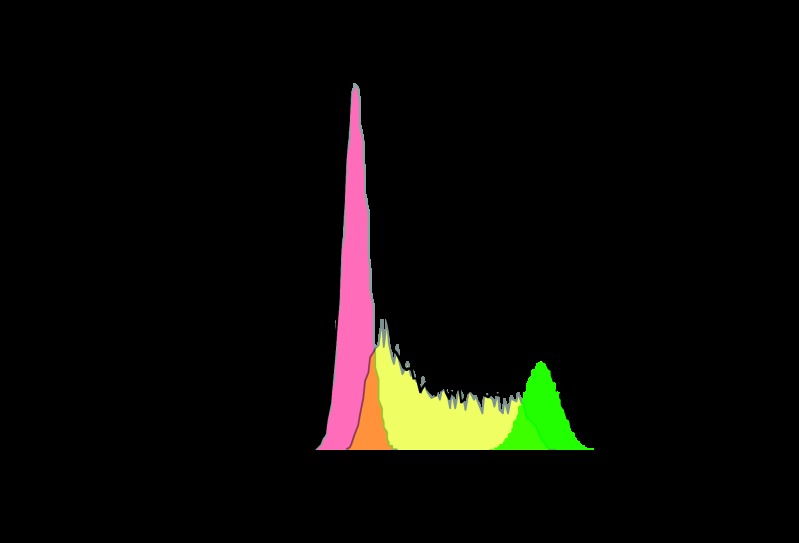
Fluorescent molecules such as propidium iodide (PI) and 7-amino-actinomycin D (7-AAD) were found to provide suitable chemical characteristics for univariate analysis of cellular DNA content by flow cytometry (Table 1) (88). PI and 7-AAD exhibit a high affinity for nucleic acids and red and far-red fluorescence, respectively. For use in cell cycle analysis, cells are fixed, permeabilized, and then treated with RNase prior to staining. This approach reveals distribution of cells in three clustered phases of the cycle (G0/G1, S, and G2/M) and makes it possible to detect apoptotic cells by fractional DNA content (sub-G1 populations) (88). Cells in G0/G1 exhibit roughly one-half the fluorescence as cells in G2/M, and cells in the S phase exhibit a range of fluorescence, as they synthesize DNA (Fig. 3).
Table 1.
DNA-labeling dyes and their characteristics
| Name of Stain | Cell Permeable | Excitation Maximum, nm | Emission maximum, nm |
|---|---|---|---|
| Propidium iodide | No | 536 | 617 |
| 7-AAD | No | 488 | 655 |
| DAPI | Yes | 350 | 470 |
| Draq5 | Yes | 488–647 | 665 |
| Hoescht 33258 | Yes | 352 | 461 |
| Hoescht 33342 | Yes | 350 | 461 |
| Hoescht 34580 | Yes | 392 | 440 |
| Vybrant DyeCycle Violet | Yes | 405 | 437 |
| Vybrant DyeCycle Green | Yes | 488 | 534 |
| Vybrant DyeCycle Orange | Yes | 488, 532 | 563 |
| Vybrant DyeCycle Ruby | Yes | 488, 633/5 | 686 |
7-AAD, 7-amino-actinomycin D; DAPI, 4′,6′-diamidino-2-phenylindole.
Fig. 3.
Cytometry-based DNA content assessment-HT1080 cells were incubated with DyeCycle Violet DNA stain (111) for 5 min and analyzed by flow cytometry. Populations in the G0/G1, S, and G2/M phases are shaded in pink, yellow, and green, respectively.
PI or 7-AAD can also be used in conjunction with BrdU or EdU pulsing to determine the rates of cell cycle progression (29). Pulse-chase labeling experiments are used to provide information about the kinetics of cell cycle progression during distinct phases. Cells are first pulsed with a labeled nucleotide analog for a duration of time, and then cells are fixed and assayed for nucleotide incorporation (new DNA synthesis) and DNA content (88). Since cells typically cycle heterogeneously, use of these dyes to study specific transitions within the cell cycle often requires cell synchronization prior to assessment for DNA content (38).
Immunohistochemical markers of cell proliferation.
A number of cell cycle pathway-associated proteins are used as immunohistochemical markers of cellular proliferation (Table 2). The most commonly used markers are Ki67, PCNA (29), and phospho-histone H3 (Ser10). Ki67 is a 360-kDa nuclear protein that can be used to detect and quantify proliferating cells (89). It is induced when quiescent, arrested cells enter the late G1-S transition (36) and continues to be expressed through the G2 and M phases. Its expression is undetectable in cells in G0 (26). Ki67 expression is elevated in a variety of human tumor tissues and is a diagnostic marker that inversely correlates with survival rates in a variety of cancers (15, 48, 89). PCNA is a δ-DNA polymerase cofactor involved in DNA replication and DNA repair (13, 36). A variety of commercially available phosphospecific antibodies for cell cycle pathway members can be used to indicate whether these proteins contain activating or inactivating phosphorylation states (e.g., phospho-Ser RB).
Table 2.
Immunohistochemical markers of cell proliferation
| Protein Name | Gene Name | Cellular Compartment | Labeled Phase |
|---|---|---|---|
| Ki67 | MKI67 | Nucleus | G1, S, G2, M |
| Proliferating cell nuclear antigen | PCNA | Nucleus | S |
| Aurora kinase A | AURKA | Nucleus | G2, M |
| Aurora kinase B | AURKB | Nucleus | G2, M |
| Phospho H3 (Ser10) histone | H3 | Nucleus | M |
| Phospho-RB (Ser807–811) | RB | Nucleus, cytosol | G1-S transition |
RB, retinoblastoma.
Biochemical assays for cell cycle assessment.
CDK activity can be measured after immunoprecipitation of specific CDKs from cellular or tissue lysates or with recombinant purified kinases. CDK kinase activity is quantified by transfer of radiolabeled phosphate from [γ-32P]ATP or by immunoblotting using phosphospecific antibodies for CDK substrates (e.g., RB, vimentin, nuclear protein in the AT region (NPAT)) (10). Direct CDK inhibitors such as flavopiridol UCN-01, paullones, hymenialdisine, and roscovitine can be incorporated into such assays to validate kinase specificity (98). Cells can be synchronized in a specific phase of the cell cycle by inducing a block at a specific checkpoint and then releasing the block and subsequently assaying as a function of time (38). Serum starvation is one way to arrest and synchronize cells in culture. In addition, a variety of compounds can be used to synchronize cells in a specific phase of the cell cycle (e.g., double-thymidine block to arrest cells in G1 and nocodazole to arrest cells in G2) (38). G1 CDKs (e.g., CDK4/6) and G2/M kinases can efficiently phosphorylate RB and histone H1 in vitro, respectively. However, cross-reactivity at lower efficiency between G1 and G2 kinases and the aforementioned substrates can be observed (10, 12).
A second biochemical approach to assess for cell cycle activity exploits the functional requirement of specific transcription factor families during cell cycle progression (e.g., E2F family members) (7). Electric mobility shift assay is a sensitive method for detecting transcription factor DNA interactions. Cell lysates, either whole cell or nuclear, are incubated with double-stranded labeled oligonucleotides (e.g., 32P or biotin end-labeled) that contain DNA binding sequences for specific transcription factors. Samples are then resolved by nondenaturing polyacrylamide gel electrophoresis. Transcription factor activity is reflected in a higher-molecular-weight shift in the migration of labeled DNA oligonucleotides. Specificity of binding is confirmed by several controls, including competition with unlabeled nucleotides, and, if possible, “supershifting” by preincubation with antibodies raised against specific transcription factors (7). Electric mobility shift assays have been used to detect E2F family activity during the G1-S transition (68) and for Foxo family members that are transiently activated during G2/M transition (e.g., FoxM1) (56, 84).
Luciferase reporters to assess for cell cycle regulation.
Luciferase assays have gained popularity due to their relative simplicity and applicability to high-throughput assays. Such assays are conducted by fusing a cell cycle-responsive promoter upstream of a destabilized firefly luciferase gene (Photinus pyralis) in an expression vector. Cells are transfected and synchronized at a specific phase of the cell cycle and then released, treated, lysed, and assessed for luminescence using a luminometer. Signals must then be normalized. A variety of methods for doing so include the use of untransfected cells, cells transfected with an empty luciferase vector, and cells transfected with a constitutive promoter not regulated by the cell cycle [e.g., EF1α (122)].
The dual-luciferase normalization method consists of cotransfecting cells with a control plasmid containing a ubiquitous promoter (e.g., phosphoglycerate kinase 1) upstream of Renilla luciferase (Renilla reniformis). The firefly luciferase signal is measured and quenched, and Renilla activity is then assessed in the same lysates. This is the most efficient method to achieve signal normalization; efficient application of this method is facilitated by use of a luminometer with built-in injectors. This promoter-based system can also be adapted to assess for cell cycle activity in living cells by switching the reporter gene to a fluorescent gene (e.g., GFP, as discussed below) or by using cell-permeable luciferase gene product substrates (32). Luciferase reporter studies are possible in live cells while controlled tissue culture growth conditions are maintained, but they require specialized laboratory equipment (e.g., LumiCycle from Actimetrix) and culture media.
Cell cycle-responsive promoters have been successfully utilized with the luciferase system to assess for cell cycle progression. The ∼100-bp (−89/+11) proximal promoter of the human cyclin A gene is repressed in G1 phase and induced upon S phase entry [as shown in NIH 3T3 cells (97)]. The 1.5-kb proximal promoter of the E2f1 gene is repressed in G0 and early G1 and is induced during the G1-S transition in REF-52 cells (47) or U2OS cells (32). Six forkhead transcription factor binding sites can drive luciferase expression during G2/M in U20S cells (32, 60).
Methods for Cell Cycle Assessment in Living Cells and Tissue
Cell-permeable dyes for live cell labeling of DNA content and cell proliferation.
A variety of membrane-permeable dyes have been developed to determine DNA content in living cells (Table 1) (70, 101, 111). Optimal dye characteristics for live cell cycle assessment include cell and tissue permeability, low cytotoxicity and phototoxicity, photostability, and stochiometric specificity for DNA with minimal affinity for RNA. Also, if possible, spectral characteristics that render it compatible with other fluorescent proteins (e.g., GFP, RFP) facilitate multiparameter assessment. DNA content staining is useful for directly quantifying the number of cells in distinct phases in the same way that cell-impermeable DNA stains are used, as described above. However, they have the added advantage that intact live cells can loaded with the dye. This is important if one wants to quantify changes in DNA content in vivo after a perturbation within a defined window of time (e.g., drug dose, injury). Alternatively, these dyes can be used to purify or quantify cells in distinct phases of the cell cycle or containing different amounts of DNA ploidy (e.g., G0/G1, S, 2N, 4N, etc.) while maintaining cell integrity for downstream applications (e.g., quantifying specific transcripts or proteins by methods that would be sensitive to cell permeabilization, e.g., high-content sequencing analysis).
Of the commercially available cell-permeable DNA dyes, 4′,6′-diamidino-2-phenylindole (DAPI), Draq5 {1,5-bis[2-(methylamino)ethyl]amino-4,8-dihydroxyanthracene-9,10-dione}, Hoescht (bis-benzimides), and Vybrant DyeCycle dyes have characteristics that include sufficient affinity and permeability to facilitate supravital staining and cytometry-based assessment of cell cycle phase distribution. DAPI (21) and the Hoescht dyes were the first dyes used in living cells because of their cell permeability characteristics and their specificity for DNA. DAPI and Hoescht dyes are excited in the UV range (350–390 nm) and emit fluorescence in the blue range. They bind strongly to the minor groove of DNA, have fairly broad emission spectra, increase fluorescence after binding to DNA, and favor AT-rich sequences. Two Hoescht stains (33258 and 33342) are used for DNA labeling and cell cycle analysis (61, 62). Hoescht stains are also substrates for the ATP-binding cassette transporter that is highly expressed on stem and progenitor cells, a property that is exploited for the use of these dyes to identify and purify stem cell populations from adult tissues (111). In general, chronic exposure to DNA-intercalating dyes can have a variety of negative effects on normal cell function, including adverse effects on cellular proliferation (100).
The spectral characteristics of DAPI and the Hoechst stains require that they be excited by UV light, thus limiting their use in long-term live cell imaging studies. Prolonged exposure of living tissue to UV light causes photobleaching and DNA damage responses that alter cell cycle progression and induce apoptosis. Moreover, establishing stable levels of nuclear dye retention for long-term imaging can be difficult and often requires repetitive loading. Continuous imaging of Hoechst-stained living cells can result in rapid cell death (70). Moreover, although UV light sources are common on microscopes, this is not the case for flow cytometers, and retrofitting a flow cytometer with UV light and detectors can be expensive. In light of these limitations, the DyeCycle Violet stain was first developed and was based on Hoechst 33342 structural characteristics. It exhibits a right-shifted excitation and a broader emission spectra than does Hoechst 33342 (Table 1) (111). Similarly to the Hoechst stain, DyeCycle Violet significantly increases fluorescence after binding to DNA. Other DyeCycle stains that emit in the green, orange, and red wavelengths that are excitable with 488-nm lasers have been developed. These dyes have a proprietary structure (Invitrogen) but are reported to be useful for sorting cells by flow cytometry such that the sorted cells can be cultured after incubation with the dye.
DRAQ5 is a deep red fluorescing bisalkylaminoanthraquinone (Ex 646, Em 681) capable of reporting three-dimensional nuclear structure and location in live cells, even in the presence of green fluorescent protein (GFP) (70, 101). Anthraquinones are synthetic DNA-binding agents that are structurally related to DNA-intercalating anthracycline antibiotics. The far-red emission spectrum of DRAQ5 facilitates its use in conjunction with commonly used green- and red-emitting fluorophores and fluorescent proteins. It stains cellular nuclei and penetrates into tissue on the second-to-minute timescale. It is cytotoxic, with 10% survival of cells exposed at 10 nM for 24 h, and thus it is not recommended for live cell sorting, where long-term survival is desired (70, 101).
Carboxyfluorescein diacetate succinimidyl ester can be used to estimate the number of times that cells have divided (67). Carboxyfluorescein diacetate succinimidyl ester passively diffuses into cells and is cleaved by intracellular esterases to produce a highly fluorescent carboxyfluorescein succinimidyl ester. The succinimidyl ester reacts with intracellular amines, forming fluorescent conjugates that are retained in daughter cells after cellular division. Each daughter cell exhibits approximately one-half of the fluorescent signal as the parent cell.
Cell cycle-responsive promoters for sensing cell cycle entry in live cells.
As mentioned above, cell cycle-dependent promoters have enabled the application of the luciferase system to assess cell cycle regulation. These promoters can also be used to drive expression of fluorescent genes to assess regulation of the cell cycle in living cells. Our laboratory generated, cloned, and characterized the 1.5-kb proximal promoter of the human Ki67 gene and used it to drive enhanced GFP expression (Ki67p-GFP; Fig. 4) (122), hypothesizing that it would be able to distinguish between quiescent and cycling cells.
Fig. 4.

Genetic reporters for cell cycle assessment in live cells and tissue; nucleus (n), cytoplasm (c), promoter (p), chicken β-actin promoter (CCAG), endogenous rosa 26 promoter (R26), and floxed PGK-neomycin resistance gene-poly A cassette (PNpA), green fluorescent protein (GFP).
The Ki67p-GFP reporter has been used to distinguish between subpopulations of cells that are arrested in G0 from those that are actively transitioning through it (27). We reported that Ki67p-GFP colocalizes with endogenous Ki67 and can be used to label proliferating cells in three-dimensional tissue cultures (122). It has the added advantage that in quiescent cells its activity is attenuated, and thus it could be used to indirectly quantify the number of cells that have exited the cell cycle specifically by determining the number of Ki67p-GFP cells that are GFP−.
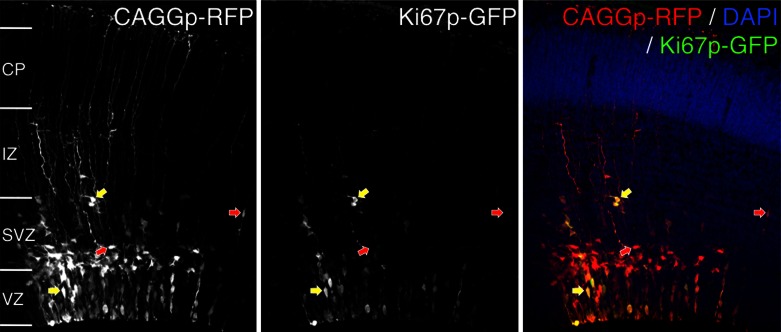
The labeling of proliferating cells with Ki67p-GFP may aid in the study of the cell proliferation in the nervous system. Recent evidence suggests that neurons generated in the same time window preferentially form connections with one another (22). To assess the ability of Ki67p-GFP to label proliferating subpopulations of highly interconnected neurons in vivo, we electroporated the developing cerebral cortex of an embryonic 14.5-day embryo in utero with Ki67p-GFP and a constitutively active chicken β-actin promoter (90) driving RFP (CAGGp-RFP) reporter constructs. Fluorescence was assessed after 24 h (Fig. 5). Ki67p-GFP labeled fewer cells than did the CAGGp-RFP construct, suggesting that low numbers of cells are actively proliferating. Moreover, in the subventricular zone, cell bodies labeled positive for Ki67p-GFP colocalized as pairs, suggesting that they are in the late stages of the M phase or have recently undergone cytokinesis (Fig. 5).
Fig. 5.
Ki67p-GFP expression in the developing nervous system. The developing cerebral cortex of an embryonic mouse was electroporated in utero with a plasmid containing the 1.5-kb proximal promoter of the human Ki67 gene upstream of GFP (Ki67p-GFP) (122) and a ubiquitously expressed reporter, CAGGp-RFP (90), at embryonic day 14.5 and fixed after 24 h. The ventricular (VZ) and subventricular zones (SVZ) contain neural precursors. These precursors produce postmitotic neurons that will migrate through the intermediate zone (IZ) to the cortical plate (CP) via the radial glial fibers projecting to the pial surface. Arrows indicate presumptive postmitotic neurons (red arrows) and mitotically active precursors (yellow arrows).
In rapidly cycling immortalized cells that are constantly reentering the G1-S transition, a limitation of using GFP as a reporter protein for cell cycle studies is its long half-life (∼26 h) (19). Thus, we attempted to use the Ki67p to drive expression of a destabilized GFP (66), but this resulted in very dim fluorescence that was inadequate for labeling proliferating cells (unpublished observations). We anticipate that using Ki67p to drive expression of the cell cycle-destabilized proteins (discussed below) will increase the utility of the Ki67p in immortalized cell types and in vivo.
Reporters based on cell cycle-dependent changes in protein localization and/or stability.
GFP fusions with core cell cycle proteins such as polo-like kinase 1 (3), Ccnb1 (42), yeast cell cycle proteins (110), P27 (123), and histone 2B (50) helped shed new light on the dynamic macromolecular changes that occur during cell cycle progression. However, overexpression of cell cycle-related proteins that retain enzyme activity can perturb the stoichiometric balances that are required for proper cell cycle regulation and are thus not optimal for serving as live cell cycle sensors (16).
Seminal work from Stubbs and Thomas (105) and Thomas (112) exploited cell cycle-dependent changes in protein localization to develop two genetically encoded “stealth” live cell cycle sensors that label cells during the G1-S and G2-M transitions. As noted above, changes in protein localization (e.g., cytoplasmic/nuclear transport) play a major role in cell cycle regulation (86). Stubbs and Thomas (105) developed a stealth G1/S reporter construct by using the ubiquitin C constitutive promoter (90) to drive expression of a cDNA encoding a GFP fused to the 131-amino acid phosphorylation-dependent subcellular localization control domain of the helicase B gene (Helb) (Fig. 4). The HLB gene is a DNA-dependent ATPase that catalyzes the unwinding of DNA during DNA replication and repair during S phase entry (34, 109). The G1/S reporter labels nuclei of cells in G0/G1 and the early S phase. During S phase transition, the fusion protein is shuttled out of the nucleus and is only in the cytoplasm during G2/M (Fig. 4).
The stealth G2/M sensor harnesses the 949-bp proximal promoter of Ccnb1 (44) to drive expression of a cDNA encoding Ccnb1 amino acids 1–170 fused to the NH2 terminus of enhanced GFP (Fig. 4). This Ccnb1 proximal promoter is active in late S phase and drives expression of the Ccnb1-enhanced GFP fusion construct. The expressed reporter translocates from the cytoplasm to the nucleus at prophase and is degraded by the encoded Ccnb1 destruction box during mitosis (16, 105, 113). Because the Ccnb1 destruction box contains an APC/C E3 ubiquitination target domain of CCNB1, the reporter protein is stable during the S phase until metaphase. The reporter is expressed and degraded in concert with endogenous Ccnb1 but does not compete for binding to CDK since it lacks the COOH-terminal sequences that comprise the Ccnb1/CDK interaction domains. Extensive clonal selection and characterization was carried out to identify a clonal U2OS line that exhibits the appropriate signal intensity and normal cell cycle kinetics (114).
Robust application of the G1/S reporter that relies on changes in cellular localization requires an environmentally controlled confocal microscope and image-processing software to automate quantification of changes in reporter localization. Moreover, it is not known whether these constructs can be used in other commonly used model organisms (e.g., Drosophila or zebrafish). However, because advanced imaging is used to assess cell cycle reporters that are dependent on changes in protein localization, one could envision quantifying changes in other cellular structures (e.g., nuclear envelop breakdown) or labeled proteins in the context of cells in specific phases of the cell cycle.
Recently, Klochendler et al. (55) generated a transgenic mouse line containing a Ccnb1-GFP fusion construct described above (16) under control of the mouse phosphoglycerate kinase 1 promoter (Fig. 4). The expression construct had been developed and characterized previously in cultured cells (35). The transgenic mouse line was used to isolate and profile the expression of dividing and nondividing adult and juvenile liver cells. Interestingly ∼10% of CCNB1/GFP-positive cells do not traverse the S phase (as measured by BrdU incorporation), suggesting that a subpopulation of cells have altered APC/C activity (55, 76).
A similar approach of fusing cell cycle protein destabilization domains to fluorescent reporters was employed by Sakaue-Sawano et al. (93) to develop the fluorescent ubiquitination-based cell cycle indicator (Fucci). This live cell sensor consists of a duel transgenic system that labels nuclei of cells in the G0/G1 and early S phases with the fast-folding monomeric Kusabira Orange (mKO2) fluorescent protein (51) and cells in late S/G2 and early M phases with a monomeric version of the green fluorescent protein Azami Green (mAg) (52). Cells in the S phase are labeled by both proteins (Fig. 4).
The mKO2 reporter is a fusion of amino acids 30–120 of the human Cdt1 protein to the carboxyl terminus of mKO2 [mKO2-hCdt1(30–120)]. Amino acids 30–120 contain a Cy motif that is targeted by SKP2 E3 ligase, a ligase that ubiquitinates a variety of cell cycle proteins during S/G2, targeting them for proteolysis (76, 108). As mentioned above, Cdt1 is a ubiquitin ligase that is involved in origin licensing and the formation of the prereplication complex during DNA replication (17, 120).
The late S and G2/M Fucci reporter fuses residues 1–110 of the human geminin (Gem) protein to the carboxyl terminus of mAG [mAG-hGem(1–110)]. Gem inhibits DNA replication during the late S and G2 phases by binding to and inhibiting Cdt1 activity to prevent incorporation of minichromosome maintenance proteins into the prereplication complex (120). It is degraded during the metaphase-anaphase transition (73). The Gem domain encoded within residues 1–110 is ubiquitinated by APC/C during late M/G1 phase, targeting it for degradation in a manner similar to that described above for CCNB1. This activity results in nuclear accumulation of mAG-hGem(1–110) during the late S and G2/M phases (Fig. 4) (93). Domains from the zebrafish orthologs of Gem and Cdt1 were used to generate a zebrafish Fucci system that has been used to generate exceptionally detailed movies of cell cycle progression in developing zebrafish embryos (106). The system has also been adapted to Drosophila (75) and Ciona (80).
CAG promoter-driven Fucci cassettes (78) were used to develop transgenic mice (93). Double-Fucci transgenic mice were generated in which every somatic cell nucleus in the developing embryo exhibits either red or green fluorescence. Fucci2 mice have been developed recently using mCherry-hCdt(30/120) and mVenus-hGem(1/110) fluorescent chimeras that provide better color contrast compared with the first-generation Fucci reporters (Fig. 4) (1). Fucci2 transgenes have also been targeted to the Rosa26 locus, thereby reducing in vivo variability caused by the CAG promoter and transgenic insertion events. Moreover, the targeting construct was developed to enable cell type-specific Fucci2 expression by Cre-mediated loxP recombination (1).
The Fucci system has greatly facilitated studies of spatial and temporal cell cycle regulation in vitro and in vivo. Fucci-expressing cells have been used to examine the complex relationships between cell cycle kinetics and fundamental cellular processes such as signaling (93), differentiation (14), cell size (103), protein compartmentation (96), and heterogeneity of tumor cell responses to anti-cancer agents (92). In the latter study, individual cells were tracked with the reporter system within a population of unsynchronized cells, and heterogeneous effects of drug doses were quantified at the single-cell level.
The Fucci system has proven to be an invaluable tool for in vivo applications such as tumor and xenograft models, where its activity as a readout of tumor cell proliferation can be tracked in intact animals using whole animal imaging (20, 93). It has also been used for advanced imaging of cell proliferation in the neural system in clarified intact brain tissue (37) and the proliferation of astrocyte populations in postnatal brains (30), among others.
As with any genetically encoded sensors, limitations of gene transfer can be significant. Many cell types, both primary and immortalized, can be difficult to transfect. In light of this, both the Ki67p-GFP and Fucci reporter constructs have been cloned into lenti-viral vectors, thereby facilitating their delivery into difficult-to-transfect and nondividing cell types. Generation of replication-deficient lenti-viral particles is straightforward when a suitable packaging cell line (e.g., HEK-293FT) and packaging vector plasmids are available. Moreover, isolation of Fucci- and Fucci2-expressing cells from Fucci transgenic mice circumvents the need for gene transfer into primary isolated cell types.
Summary and Future Perspectives
Cell cycle progression is a fundamental process in all living organisms. Classically, this process has been studied using immortalized cell lines and/or approaches that require tissue fixation, thereby limiting our understanding of important features of cell cycle progression, including its heterogenous nature. In vivo, the cell cycle is dynamically and tightly regulated by multiple mechanisms and signaling pathways that receive inputs that vary depending on cell type and cellular niche.
This review highlights a number of classic and emerging techniques for cell cycle assessment. These emerging techniques will likely shed light on new genes and proteins that can be targeted to block or induce cell cycle progression in specific cell populations for experimental studies and therapeutic purposes. For example, the cell cycle pathway is a primary target in cancer therapy. However, tumor cell heterogeneity contributes to cancer dormancy, a process that is poorly understood. Cancer dormancy occurs when residual disease is present but the patient remains asymptomatic. This process is common in many cancers [e.g., 20–45% of patients with breast or prostate cancer will relapse years or decades later (23, 53, 85)].
The transition of a cancer cell into cellular dormancy (cell cycle arrest or cellular quiescence) is one of several proposed mechanisms that cause cancer dormancy (2). Live cell cycle sensors that enable single-cell resolution of cell cycle kinetics in vivo over extended times (perhaps even on the order of months to years) are likely to increase our understanding of this important but poorly understood process and will hopefully aid in the discovery of new therapeutics in this area.
Conversely, the process of inducing cell cycle progression in other therapeutic settings (e.g., during cell therapy for regenerative medicine) is also a promising concept, assuming this process can be tightly controlled. For example, cardiac disorders are a leading cause of death and disability worldwide, in part as a consequence of the limited capacity for adult cardiac myocytes (CMs) to reenter the cell cycle and proliferate. Cardiac ischemia, in particular as a consequence of myocardial infarction, can destroy CMs and, in addition, other cell types essential for a normal functioning myocardium (e.g., endothelial and smooth muscle cells in the vasculature).
Although significant progress has been made in promoting the differentiation of embryonic and adult stem cells into terminally committed lineages (e.g., CMs), far less emphasis has been placed on targeting pathways to increase the proliferative expansion of progenitor cells for therapeutic applications. Conceptually, such cells could be of greater therapeutic benefit if they could be transplanted in sufficient quantities, by controlling their proliferative expansion in vitro or in vivo, and then guided to differentiate into multiple cell types in the target organ. We anticipate that these and other newly developed cell cycle reporter systems will enable studies in these areas.
GRANTS
D. S. Bortone was supported by an National Institutes of Health (NIH) Research Service Award Grant (National Institute of Neurological Disorders and Stroke: 1-F32-NS-076185-01A1). A. C. Zambon was supported by an American Heart Association Grant (10SDG2630130) and NIH Grants 1-U54-HL-108460, 8-UL1-TR-000100, and P01-HL-098053. L. Henderson was supported by National Institute of General Medical Sciences Grant 2-R25-GM-083275.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
L.H., C.L., and A.C.Z. contributed to conception and design of the research; L.H., D.S.B., and A.C.Z. analyzed the data; L.H., D.S.B., C.L., and A.C.Z. prepared the figures; L.H. and A.C.Z. drafted the manuscript; L.H., D.S.B., C.L., and A.C.Z. edited and revised the manuscript; L.H., D.S.B., C.L., and A.C.Z. approved the final version of the manuscript; D.S.B. and A.C.Z. performed the experiments; D.S.B. and A.C.Z. interpreted the results of the experiments.
ACKNOWLEDGMENTS
We thank the University of California San Diego Neuroscience Microscopy Facility (P30 NS047101) for the use of their imaging equipment.
REFERENCES
- 1. Abe T, Sakaue-Sawano A, Kiyonari H, Shioi G, Inoue K, Horiuchi T, Nakao K, Miyawaki A, Aizawa S, Fujimori T. Visualization of cell cycle in mouse embryos with Fucci2 reporter directed by Rosa26 promoter. Development 140: 237–246, 2013 [DOI] [PubMed] [Google Scholar]
- 2. Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer 7: 834–846, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arnaud L, Pines J, Nigg EA. GFP tagging reveals human Polo-like kinase 1 at the kinetochore/centromere region of mitotic chromosomes. Chromosoma 107: 424–429, 1998 [DOI] [PubMed] [Google Scholar]
- 4. Basto R, Pines J. The centrosome opens the way to mitosis. Dev Cell 12: 475–477, 2007 [DOI] [PubMed] [Google Scholar]
- 5. Bauer S, Patterson PH. The cell cycle-apoptosis connection revisited in the adult brain. J Cell Biol 171: 641–650, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bernardin A, Cazet A, Guyon L, Delannoy P, Vinet F, Bonnaffe D, Texier I. Copper-free click chemistry for highly luminescent quantum dot conjugates: application to in vivo metabolic imaging. Bioconjug Chem 21: 583–588, 2010 [DOI] [PubMed] [Google Scholar]
- 7. Bicknell KA. Forkhead (FOX) transcription factors and the cell cycle: measurement of DNA binding by FoxO and FoxM transcription factors. Methods Mol Biol 296: 247–262, 2005 [DOI] [PubMed] [Google Scholar]
- 8. Blangy A, Lane HA, d'Hérin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83: 1159–1169, 1995 [DOI] [PubMed] [Google Scholar]
- 9. Borgs L, Beukelaers P, Vandenbosch R, Belachew S, Nguyen L, Malgrange B. Cell “circadian” cycle: new role for mammalian core clock genes. Cell Cycle 8: 832–837, 2009 [DOI] [PubMed] [Google Scholar]
- 10. Brooks G. Cyclins, cyclin-dependent kinases, and cyclin-dependent kinase inhibitors: detection methods and activity measurements. Methods Mol Biol 296: 291–298, 2005 [DOI] [PubMed] [Google Scholar]
- 11. Brooks G, Poolman RA, Li JM. Arresting developments in the cardiac myocyte cell cycle: role of cyclin-dependent kinase inhibitors. Cardiovasc Res 39: 301–311, 1998 [DOI] [PubMed] [Google Scholar]
- 12. Brooks G, Poolman RA, McGill CJ, Li JM. Expression and activities of cyclins and cyclin-dependent kinases in developing rat ventricular myocytes. J Mol Cell Cardiol 29: 2261–2271, 1997 [DOI] [PubMed] [Google Scholar]
- 13. Burkovics P, Hajdu I, Szukacsov V, Unk I, Haracska L. Role of PCNA-dependent stimulation of 3′-phosphodiesterase and 3′-5′ exonuclease activities of human Ape2 in repair of oxidative DNA damage. Nucleic Acids Res 37: 4247–4255, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Calder A, Roth-Albin I, Bhatia S, Pilquil C, Lee JH, Bhatia M, Levadoux-Martin M, McNicol J, Russell J, Collins T, Draper JS. Lengthened G1 phase indicates differentiation status in human embryonic stem cells. Stem Cells Dev 22: 279–295, 2013 [DOI] [PubMed] [Google Scholar]
- 15. Chen L, Li X, Wang GL, Wang Y, Zhu YY, Zhu J. Clinicopathological significance of overexpression of TSPAN1, Ki67 and CD34 in gastric carcinoma. Tumori 94: 531–538, 2008 [DOI] [PubMed] [Google Scholar]
- 16. Clute P, Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat Cell Biol 1: 82–87, 1999 [DOI] [PubMed] [Google Scholar]
- 17. Cook JG, Chasse DA, Nevins JR. The regulated association of Cdt1 with minichromosome maintenance proteins and Cdc6 in mammalian cells. J Biol Chem 279: 9625–9633, 2004 [DOI] [PubMed] [Google Scholar]
- 18. Coqueret O. New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol 13: 65–70, 2003 [DOI] [PubMed] [Google Scholar]
- 19. Corish P, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng 12: 1035–1040, 1999 [DOI] [PubMed] [Google Scholar]
- 20. Dan S, Okamura M, Mukai Y, Yoshimi H, Inoue Y, Hanyu A, Sakaue-Sawano A, Imamura T, Miyawaki A, Yamori T. ZSTK474, a specific phosphatidylinositol 3-kinase inhibitor, induces G1 arrest of the cell cycle in vivo. Eur J Cancer 48: 936–943, 2012 [DOI] [PubMed] [Google Scholar]
- 21. Darzynkiewicz Z, Williamson B, Carswell EA, Old LJ. Cell cycle-specific effects of tumor necrosis factor. Cancer Res 44: 83–90, 1984 [PubMed] [Google Scholar]
- 22. Deguchi Y, Donato F, Galimberti I, Cabuy E, Caroni P. Temporally matched subpopulations of selectively interconnected principal neurons in the hippocampus. Nat Neurosci 14: 495–504, 2011 [DOI] [PubMed] [Google Scholar]
- 23. Demicheli R, Fornili M, Ambrogi F, Higgins K, Boyd JA, Biganzoli E, Kelsey CR. Recurrence dynamics for non-small-cell lung cancer: effect of surgery on the development of metastases. J Thorac Oncol 7: 723–730, 2012 [DOI] [PubMed] [Google Scholar]
- 24. Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev 12: 2245–2262, 1998 [DOI] [PubMed] [Google Scholar]
- 25. Ebelt H, Hufnagel N, Neuhaus P, Neuhaus H, Gajawada P, Simm A, Muller-Werdan U, Werdan K, Braun T. Divergent siblings: E2F2 and E2F4 but not E2F1 and E2F3 induce DNA synthesis in cardiomyocytes without activation of apoptosis. Circ Res 96: 509–517, 2005 [DOI] [PubMed] [Google Scholar]
- 26. Endl E, Steinbach P, Knuchel R, Hofstadter F. Analysis of cell cycle-related Ki-67 and p120 expression by flow cytometric BrdUrd-Hoechst/7AAD and immunolabeling technique. Cytometry 29: 233–241, 1997 [PubMed] [Google Scholar]
- 27. Fadeev RS, Chekanov AV, Dolgikh NV, Akatov VS. [Increase in resistance of A431 cancer cells to TRAIL-induced apoptosis in confluent cultures]. Biofizika 57: 649–654, 2012 [PubMed] [Google Scholar]
- 28. Felfly H, Xue J, Zambon AC, Muotri A, Zhou D, Haddad GG. Identification of a neuronal gene expression signature: role of cell cycle arrest in murine neuronal differentiation in vitro. Am J Physiol Regul Integr Comp Physiol 301: R727–R745, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gatti G, Maresca G, Natoli M, Florenzano F, Nicolin A, Felsani A, D'Agnano I. MYC prevents apoptosis and enhances endoreduplication induced by paclitaxel. PLoS One 4: e5442, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ge WP, Miyawaki A, Gage FH, Jan YN, Jan LY. Local generation of glia is a major astrocyte source in postnatal cortex. Nature 484: 376–380, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature 349: 132–138, 1991 [DOI] [PubMed] [Google Scholar]
- 32. Grant GD, Gamsby J, Martyanov V, Brooks L, 3rd, George LK, Mahoney JM, Loros JJ, Dunlap JC, Whitfield ML. Live-cell monitoring of periodic gene expression in synchronous human cells identifies Forkhead genes involved in cell cycle control. Mol Biol Cell 23: 3079–3093, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Greene LA, Biswas SC, Liu DX. Cell cycle molecules and vertebrate neuron death: E2F at the hub. Cell Death Differ 11: 49–60, 2004 [DOI] [PubMed] [Google Scholar]
- 34. Guler GD, Liu H, Vaithiyalingam S, Arnett DR, Kremmer E, Chazin WJ, Fanning E. Human DNA helicase B (HDHB) binds to replication protein A and facilitates cellular recovery from replication stress. J Biol Chem 287: 6469–6481, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hagting A, Karlsson C, Clute P, Jackman M, Pines J. MPF localization is controlled by nuclear export. EMBO J 17: 4127–4138, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hall PA, Woods AL. Immunohistochemical markers of cellular proliferation: achievements, problems and prospects. Cell Tissue Kinet 23: 505–522, 1990 [DOI] [PubMed] [Google Scholar]
- 37. Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci 14: 1481–1488, 2011 [DOI] [PubMed] [Google Scholar]
- 38. Harper JV. Synchronization of cell populations in G1/S and G2/M phases of the cell cycle. Methods Mol Biol 296: 157–166, 2005 [DOI] [PubMed] [Google Scholar]
- 39. Healy S, Khan P, He S, Davie JR. Histone H3 phosphorylation, immediate-early gene expression, and the nucleosomal response: a historical perspective. Biochem Cell Biol 90: 39–54, 2012 [DOI] [PubMed] [Google Scholar]
- 40. Herbig U, Griffith JW, Fanning E. Mutation of cyclin/cdk phosphorylation sites in HsCdc6 disrupts a late step in initiation of DNA replication in human cells. Mol Biol Cell 11: 4117–4130, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hinchcliffe EH, Sluder G. “It takes two to tango”: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev 15: 1167–1181, 2001 [DOI] [PubMed] [Google Scholar]
- 42. Huang J, Raff JW. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J 18: 2184–2195, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Humphrey T, Brooks G. Cell Cycle Control Mechanisms and Protocols. Totowa, NJ: Humana, 2005, p. 402. [Google Scholar]
- 44. Hwang A, Maity A, McKenna WG, Muschel RJ. Cell cycle-dependent regulation of the cyclin B1 promoter. J Biol Chem 270: 28419–28424, 1995 [DOI] [PubMed] [Google Scholar]
- 45. Izumi T, Maller JL. Elimination of cdc2 phosphorylation sites in the cdc25 phosphatase blocks initiation of M-phase. Mol Biol Cell 4: 1337–1350, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jiang W, McDonald D, Hope TJ, Hunter T. Mammalian Cdc7-Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J 18: 5703–5713, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Johnson DG, Ohtani K, Nevins JR. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev 8: 1514–1525, 1994 [DOI] [PubMed] [Google Scholar]
- 48. Jones RL, Salter J, A'Hern R, Nerurkar A, Parton M, Reis-Filho JS, Smith IE, Dowsett M. The prognostic significance of Ki67 before and after neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat 2008 [DOI] [PubMed] [Google Scholar]
- 49. Jordan SN, Canman JC. Rho GTPases in animal cell cytokinesis: an occupation by the one percent. Cytoskeleton (Hoboken) 69: 919–930, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol 8: 377–385, 1998 [DOI] [PubMed] [Google Scholar]
- 51. Karasawa S, Araki T, Nagai T, Mizuno H, Miyawaki A. Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem J 381: 307–312, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Karasawa S, Araki T, Yamamoto-Hino M, Miyawaki A. A green-emitting fluorescent protein from Galaxeidae coral and its monomeric version for use in fluorescent labeling. J Biol Chem 278: 34167–34171, 2003 [DOI] [PubMed] [Google Scholar]
- 53. Karrison TG, Ferguson DJ, Meier P. Dormancy of mammary carcinoma after mastectomy. J Natl Cancer Inst 91: 80–85, 1999 [DOI] [PubMed] [Google Scholar]
- 54. Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature 432: 316–323, 2004 [DOI] [PubMed] [Google Scholar]
- 55. Klochendler A, Weinberg-Corem N, Moran M, Swisa A, Pochet N, Savova V, Vikesa J, Van de Peer Y, Brandeis M, Regev A, Nielsen FC, Dor Y, Eden A. A transgenic mouse marking live replicating cells reveals in vivo transcriptional program of proliferation. Dev Cell 23: 681–690, 2012 [DOI] [PubMed] [Google Scholar]
- 56. Korver W, Roose J, Clevers H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res 25: 1715–1719, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kuan CY, Schloemer AJ, Lu A, Burns KA, Weng WL, Williams MT, Strauss KI, Vorhees CV, Flavell RA, Davis RJ, Sharp FR, Rakic P. Hypoxia-ischemia induces DNA synthesis without cell proliferation in dying neurons in adult rodent brain. J Neurosci 24: 10763–10772, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes Dev 11: 847–862, 1997 [DOI] [PubMed] [Google Scholar]
- 59. Laflamme MA, Murry CE. Heart regeneration. Nature 473: 326–335, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol 7: 126–136, 2005 [DOI] [PubMed] [Google Scholar]
- 61. Latt SA, Stetten G. Spectral studies on 33258 Hoechst and related bisbenzimidazole dyes useful for fluorescent detection of deoxyribonucleic acid synthesis. J Histochem Cytochem 24: 24–33, 1976 [DOI] [PubMed] [Google Scholar]
- 62. Latt SA, Stetten G, Juergens LA, Willard HF, Scher CD. Recent developments in the detection of deoxyribonucleic acid synthesis by 33258 Hoechst fluorescence. J Histochem Cytochem 23: 493–505, 1975 [DOI] [PubMed] [Google Scholar]
- 63. Lehner B, Sandner B, Marschallinger J, Lehner C, Furtner T, Couillard-Despres S, Rivera FJ, Brockhoff G, Bauer HC, Weidner N, Aigner L. The dark side of BrdU in neural stem cell biology: detrimental effects on cell cycle, differentiation and survival. Cell Tissue Res 345: 313–328, 2011 [DOI] [PubMed] [Google Scholar]
- 64. Lemieux E, Boucher MJ, Mongrain S, Boudreau F, Asselin C, Rivard N. Constitutive activation of the MEK/ERK pathway inhibits intestinal epithelial cell differentiation. Am J Physiol Gastrointest Liver Physiol 301: G719–G730, 2011 [DOI] [PubMed] [Google Scholar]
- 65. Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol 28: 1737–1746, 1996 [DOI] [PubMed] [Google Scholar]
- 66. Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR. Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem 273: 34970–34975, 1998 [DOI] [PubMed] [Google Scholar]
- 67. Lyons AB. Divided we stand: tracking cell proliferation with carboxyfluorescein diacetate succinimidyl ester. Immunol Cell Biol 77: 509–515, 1999 [DOI] [PubMed] [Google Scholar]
- 68. Ma Y, Kurtyka CA, Boyapalle S, Sung SS, Lawrence H, Guida W, Cress WD. A small-molecule E2F inhibitor blocks growth in a melanoma culture model. Cancer Res 68: 6292–6299, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mailand N, Diffley JF. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell 122: 915–926, 2005 [DOI] [PubMed] [Google Scholar]
- 70. Martin RM, Leonhardt H, Cardoso MC. DNA labeling in living cells. Cytometry A 67: 45–52, 2005 [DOI] [PubMed] [Google Scholar]
- 71. Maslov AY, Barone TA, Plunkett RJ, Pruitt SC. Neural stem cell detection, characterization, and age-related changes in the subventricular zone of mice. J Neurosci 24: 1726–1733, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Matuo R, Sousa FG, Soares DG, Bonatto D, Saffi J, Escargueil AE, Larsen AK, Henriques JA. Saccharomyces cerevisiae as a model system to study the response to anticancer agents. Cancer Chemother Pharmacol 70: 491–502, 2012 [DOI] [PubMed] [Google Scholar]
- 73. McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93: 1043–1053, 1998 [DOI] [PubMed] [Google Scholar]
- 74. Miele L. The biology of cyclins and cyclin-dependent protein kinases: an introduction. Methods Mol Biol 285: 3–21, 2004 [DOI] [PubMed] [Google Scholar]
- 75. Nakajima Y, Kuranaga E, Sugimura K, Miyawaki A, Miura M. Nonautonomous apoptosis is triggered by local cell cycle progression during epithelial replacement in Drosophila. Mol Cell Biol 31: 2499–2512, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer 6: 369–381, 2006 [DOI] [PubMed] [Google Scholar]
- 77. Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol 2: 21–32, 2001 [DOI] [PubMed] [Google Scholar]
- 78. Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108: 193–199, 1991 [DOI] [PubMed] [Google Scholar]
- 79. Nowakowski RS, Lewin SB, Miller MW. Bromodeoxyuridine immunohistochemical determination of the lengths of the cell cycle and the DNA-synthetic phase for an anatomically defined population. J Neurocytol 18: 311–318, 1989 [DOI] [PubMed] [Google Scholar]
- 80. Ogura Y, Sakaue-Sawano A, Nakagawa M, Satoh N, Miyawaki A, Sasakura Y. Coordination of mitosis and morphogenesis: role of a prolonged G2 phase during chordate neurulation. Development 138: 577–587, 2011 [DOI] [PubMed] [Google Scholar]
- 81. Paddy MR, Saumweber H, Agard DA, Sedat JW. Time-resolved, in vivo studies of mitotic spindle formation and nuclear lamina breakdown in Drosophila early embryos. J Cell Sci 109: 591–607, 1996 [DOI] [PubMed] [Google Scholar]
- 82. Pelham RJ, Chang F. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature 419: 82–86, 2002 [DOI] [PubMed] [Google Scholar]
- 83. Perry JA, Kornbluth S. Cdc25 and Wee1: analogous opposites? Cell Div 2: 12, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Petrovic V, Costa RH, Lau LF, Raychaudhuri P, Tyner AL. FoxM1 regulates growth factor-induced expression of kinase-interacting stathmin (KIS) to promote cell cycle progression. J Biol Chem 283: 453–460, 2008 [DOI] [PubMed] [Google Scholar]
- 85. Pfitzenmaier J, Ellis WJ, Arfman EW, Hawley S, McLaughlin PO, Lange PH, Vessella RL. Telomerase activity in disseminated prostate cancer cells. BJU Int 97: 1309–1313, 2006 [DOI] [PubMed] [Google Scholar]
- 86. Pines J. Four-dimensional control of the cell cycle. Nat Cell Biol 1: E73–E79, 1999 [DOI] [PubMed] [Google Scholar]
- 87. Poolman RA, Gilchrist R, Brooks G. Cell cycle profiles and expressions of p21CIP1 AND P27KIP1 during myocyte development. Int J Cardiol 67: 133–142, 1998 [DOI] [PubMed] [Google Scholar]
- 88. Pozarowski P, Darzynkiewicz Z. Analysis of cell cycle by flow cytometry. Methods Mol Biol 281: 301–311, 2004 [DOI] [PubMed] [Google Scholar]
- 89. Preusser M, Heinzl H, Gelpi E, Hoftberger R, Fischer I, Pipp I, Milenkovic I, Wohrer A, Popovici F, Wolfsberger S, Hainfellner JA. Ki67 index in intracranial ependymoma: a promising histopathological candidate biomarker. Histopathology 53: 39–47, 2008 [DOI] [PubMed] [Google Scholar]
- 90. Qin JY, Zhang L, Clift KL, Hulur I, Xiang AP, Ren BZ, Lahn BT. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS One 5: e10611, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ray A, James MK, Larochelle S, Fisher RP, Blain SW. p27Kip1 inhibits cyclin D-cyclin-dependent kinase 4 by two independent modes. Mol Cell Biol 29: 986–999, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sakaue-Sawano A, Kobayashi T, Ohtawa K, Miyawaki A. Drug-induced cell cycle modulation leading to cell-cycle arrest, nuclear mis-segregation, or endoreplication. BMC Cell Biol 12: 2, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, Imamura T, Ogawa M, Masai H, Miyawaki A. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132: 487–498, 2008 [DOI] [PubMed] [Google Scholar]
- 94. Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA 105: 2415–2420, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sangfelt O, Erickson S, Castro J, Heiden T, Gustafsson A, Einhorn S, Grandér D. Molecular mechanisms underlying interferon-alpha-induced G0/G1 arrest: CKI-mediated regulation of G1 Cdk-complexes and activation of pocket proteins. Oncogene 18: 2798–2810, 1999 [DOI] [PubMed] [Google Scholar]
- 96. Santos SD, Wollman R, Meyer T, Ferrell JE., Jr Spatial positive feedback at the onset of mitosis. Cell 149: 1500–1513, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Schulze A, Zerfass K, Spitkovsky D, Middendorp S, Berges J, Helin K, Jansen-Durr P, Henglein B. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc Natl Acad Sci USA 92: 11264–11268, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Senderowicz AM. Assays for cyclin-dependent kinase inhibitors. Methods Mol Biol 285: 69–78, 2004 [DOI] [PubMed] [Google Scholar]
- 99. Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 13: 1501–1512, 1999 [DOI] [PubMed] [Google Scholar]
- 100. Siemann DW, Keng PC. Cell cycle specific toxicity of the Hoechst 33342 stain in untreated or irradiated murine tumor cells. Cancer Res 46: 3556–3559, 1986 [PubMed] [Google Scholar]
- 101. Smith PJ, Blunt N, Wiltshire M, Hoy T, Teesdale-Spittle P, Craven MR, Watson JV, Amos WB, Errington RJ, Patterson LH. Characteristics of a novel deep red/infrared fluorescent cell-permeant DNA probe, DRAQ5, in intact human cells analyzed by flow cytometry, confocal and multiphoton microscopy. Cytometry 40: 280–291, 2000 [DOI] [PubMed] [Google Scholar]
- 102. Smits VA, Medema RH. Checking out the G(2)/M transition. Biochim Biophys Acta 1519: 1–12, 2001 [DOI] [PubMed] [Google Scholar]
- 103. Son S, Tzur A, Weng Y, Jorgensen P, Kim J, Kirschner MW, Manalis SR. Direct observation of mammalian cell growth and size regulation. Nat Methods 9: 910–912, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol Heart Circ Physiol 271: H2183–H2189, 1996 [DOI] [PubMed] [Google Scholar]
- 105. Stubbs S, Thomas N. Dynamic green fluorescent protein sensors for high-content analysis of the cell cycle. Methods Enzymol 414: 1–21, 2006 [DOI] [PubMed] [Google Scholar]
- 106. Sugiyama M, Sakaue-Sawano A, Iimura T, Fukami K, Kitaguchi T, Kawakami K, Okamoto H, Higashijima S, Miyawaki A. Illuminating cell-cycle progression in the developing zebrafish embryo. Proc Natl Acad Sci USA 106: 20812–20817, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Tamamori-Adachi M, Ito H, Sumrejkanchanakij P, Adachi S, Hiroe M, Shimizu M, Kawauchi J, Sunamori M, Marumo F, Kitajima S, Ikeda MA. Critical role of cyclin D1 nuclear import in cardiomyocyte proliferation. Circ Res 92: e12–e19, 2003 [DOI] [PubMed] [Google Scholar]
- 108. Tamamori-Adachi M, Takagi H, Hashimoto K, Goto K, Hidaka T, Koshimizu U, Yamada K, Goto I, Maejima Y, Isobe M, Nakayama KI, Inomata N, Kitajima S. Cardiomyocyte proliferation and protection against post-myocardial infarction heart failure by cyclin D1 and Skp2 ubiquitin ligase. Cardiovasc Res 80: 181–190, 2008 [DOI] [PubMed] [Google Scholar]
- 109. Taneja P, Gu J, Peng R, Carrick R, Uchiumi F, Ott RD, Gustafson E, Podust VN, Fanning E. A dominant-negative mutant of human DNA helicase B blocks the onset of chromosomal DNA replication. J Biol Chem 277: 40853–40861, 2002 [DOI] [PubMed] [Google Scholar]
- 110. Tatebe H, Goshima G, Takeda K, Nakagawa T, Kinoshita K, Yanagida M. Fission yeast living mitosis visualized by GFP-tagged gene products. Micron 32: 67–74, 2001 [DOI] [PubMed] [Google Scholar]
- 111. Telford WG, Bradford J, Godfrey W, Robey RW, Bates SE. Side population analysis using a violet-excited cell-permeable DNA binding dye. Stem Cells 25: 1029–1036, 2007 [DOI] [PubMed] [Google Scholar]
- 112. Thomas N. Lighting the circle of life: fluorescent sensors for covert surveillance of the cell cycle. Cell Cycle 2: 545–549, 2003 [PubMed] [Google Scholar]
- 113. Thomas N, Goodyer I. Stealth sensors: real time monitoring of the cell cycle. Drug Disc Today Targets 2: 26–33, 2003 [Google Scholar]
- 114. Thomas N, Kenrick M, Giesler T, Kiser G, Tinkler H, Stubbs S. Characterization and gene expression profiling of a stable cell line expressing a cell cycle GFP sensor. Cell Cycle 4: 191–195, 2005 [DOI] [PubMed] [Google Scholar]
- 115. Tuttle AH, Rankin MM, Teta M, Sartori DJ, Stein GM, Kim GJ, Virgilio C, Granger A, Zhou D, Long SH, Schiffman AB, Kushner JA. Immunofluorescent detection of two thymidine analogues (CldU and IdU) in primary tissue. J Vis Exp. 46: 2166, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Valcourt JR, Lemons JM, Haley EM, Kojima M, Demuren OO, Coller HA. Staying alive: metabolic adaptations to quiescence. Cell Cycle 11: 1680–1696, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Vega CJ, Peterson DA. Stem cell proliferative history in tissue revealed by temporal halogenated thymidine analog discrimination. Nat Methods 2: 167–169, 2005 [DOI] [PubMed] [Google Scholar]
- 118. Walsh S, Ponten A, Fleischmann BK, Jovinge S. Cardiomyocyte cell cycle control and growth estimation in vivo—an analysis based on cardiomyocyte nuclei. Cardiovasc Res 86: 365–373, 2010 [DOI] [PubMed] [Google Scholar]
- 119. Williams GH, Stoeber K. The cell cycle and cancer. J Pathol 226: 352–364, 2012 [DOI] [PubMed] [Google Scholar]
- 120. Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290: 2309–2312, 2000 [DOI] [PubMed] [Google Scholar]
- 121. Yamashiro S, Yamakita Y, Ishikawa R, Matsumura F. Mitosis-specific phosphorylation causes 83K non-muscle caldesmon to dissociate from microfilaments. Nature 344: 675–678, 1990 [DOI] [PubMed] [Google Scholar]
- 122. Zambon AC. Use of the Ki67 promoter to label cell cycle entry in living cells. Cytometry A 77: 564–570, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Zeng Y, Hirano K, Hirano M, Nishimura J, Kanaide H. Minimal requirements for the nuclear localization of p27(Kip1), a cyclin-dependent kinase inhibitor. Biochem Biophys Res Commun 274: 37–42, 2000 [DOI] [PubMed] [Google Scholar]
- 124. Zhu LL, Wu LY, Yew DT, Fan M. Effects of hypoxia on the proliferation and differentiation of NSCs. Mol Neurobiol 31: 231–242, 2005 [DOI] [PubMed] [Google Scholar]