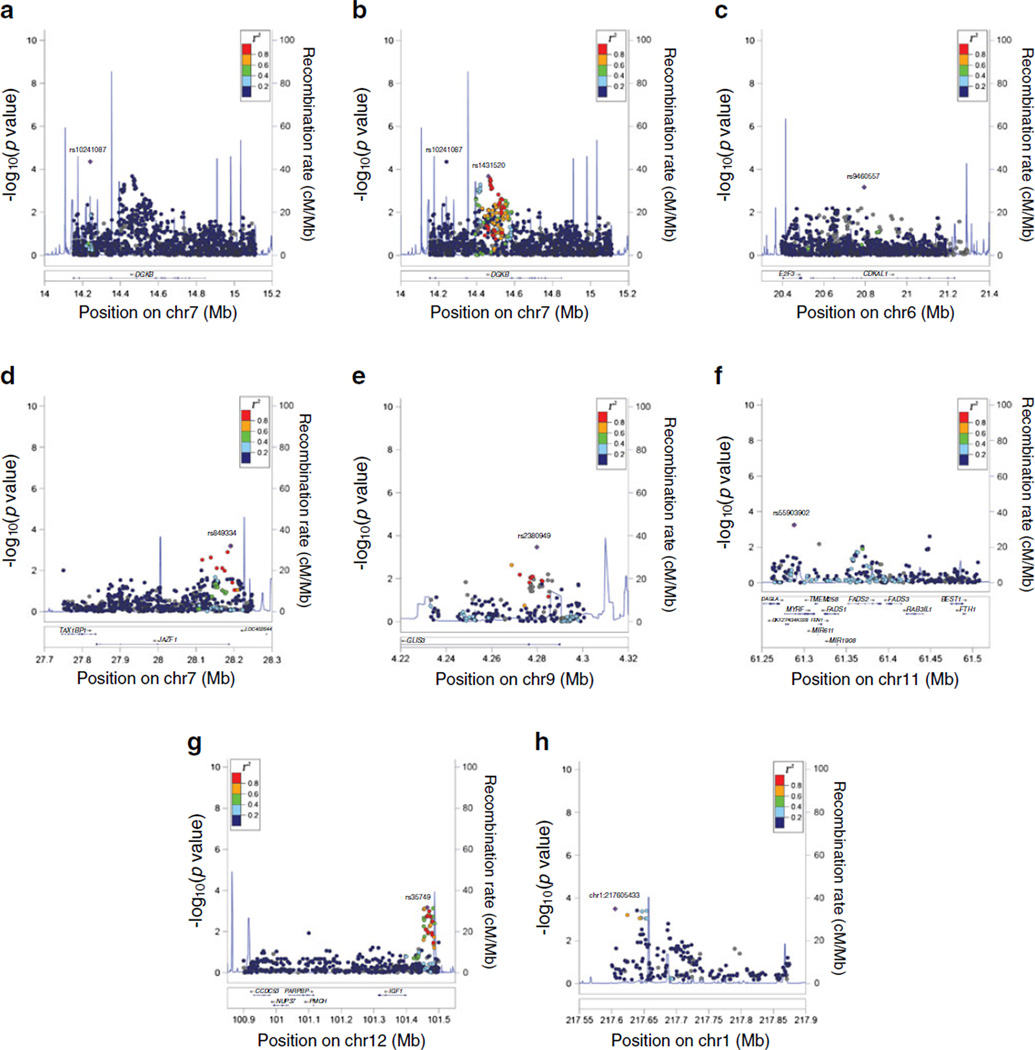
Fig. 1.
Regional association plots of association with insulin clearance. The plots display genes and recombination rates in each region. SNP associations with insulin clearance are plotted as −log10(p value). The SNP used as the index SNP in each plot is plotted as a purple diamond and labelled. Linkage disequilibrium of nearby SNPs with the index SNP is indicated by colors: dark blue for r2 < 0.2; light blue for 0.2 < r2 < 0.4; green for 0.4 < r2 < 0.6; orange for 0.6 < r2 < 0.8; and red for r2 > 0.8. (a) DGKB (based on rs10241087), (b) DGKB (based on rs1431520), (c) CDKAL1 (based on rs9460557), (d) JAZF1 (based on rs849334), (e) GLIS3 (based on rs2380949), (f) FADS1 (based on rs55903902), (g) IGF1 (based on rs35749), (h) LYPLAL1 (based on chr1:217605433)