Figure 2.
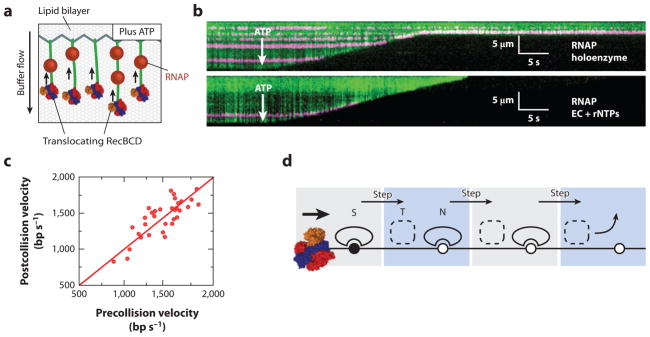
(a) An illustration of the single-molecule DNA curtain assay used to observe RecBCD-roadblock collisions. Individual DNA molecules are tethered to a fluid lipid bilayer via a biotin-streptavidin interaction and organized at nanofabricated chrome barriers. RecBCD is loaded at the free DNA ends and fluorescent RNA polymerase (RNAP) is deposited on the native promoters along the DNA. The DNA is visualized by YOYO1, an intercalating fluorescent dye that does not interfere with RecBCD activity. The reaction is initiated by supplementing the flow buffer with ATP. As the DNA is degraded by RecBCD helicase/nuclease activity, the time-dependent decrease in DNA length serves as readout of RecBCD translocation. (b) Kymographs of RecBCD pushing and evicting RNAP holoenzyme and elongation complexes (ECs) from DNA. In all kymographs, the tethered end of the DNA is at the top, the free end is at the bottom, and buffer flow is from top to bottom. RecBCD is able to efficiently push and eventually displace RNAP holoenzyme (top panel ), transcribing ECs (bottom panel ). (c) A scatter plot of pre- and postcollision RecBCD velocities. The red line is a fit to the data with a slope of 1.0. RecBCD does not change velocity upon collision with RNAP. (d ) A transition-state ejection model for roadblock displacement by RecBCD. Most roadblocks are displaced from RecBCD as they are pushed from one nonspecific DNA site to the next. Translocating RecBCD initially displaces the protein roadblock from a high-affinity specific site (S). The roadblock must pass through a much more weakly bound intermediate state (T) as it is pushed from one site to the next, followed by re-equilibration as a DNA-bound nonspecific complex (N). Subsequent steps by RecBCD continue to push the protein roadblocks from one nonspecific site to the next, until the proteins are eventually evicted from the DNA.
