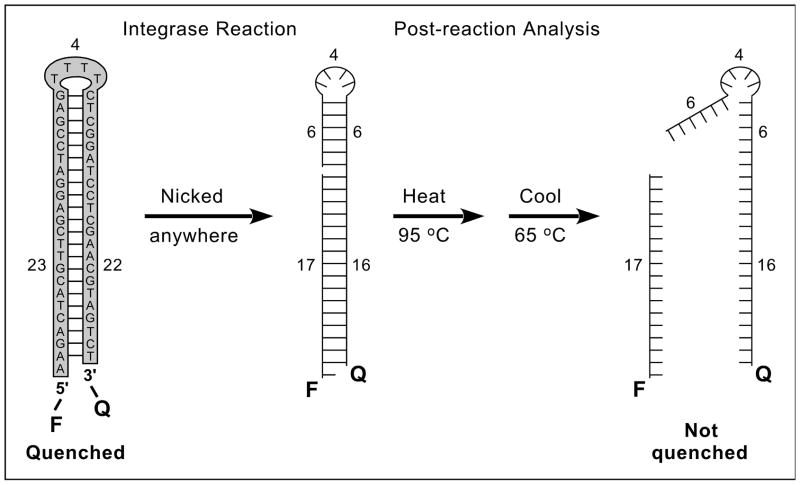
Figure 1. Nonradioactive, plate-based, solution-phase assay for nonspecific DNA nicking.
The sequence of the 49-mer oligonucleotide substrate is shown at the left, with base-pairing indicated by dashes. This DNA is 5′-labeled with a fluorophore (F), 3′-tagged with a quencher (Q), and designed to form a hairpin that brings these groups into proximity to quench fluorescence (numbers indicate lengths of structural features in nucleotides). During reactions with integrase, nicking on either side of the hairpin unlinks the F and Q groups (a nick at position 17 is used as an example, and the DNA is now shown without sequence). Completed reactions are analyzed by heating to 95°C to denature the DNA, then cooling to 65°C and measuring the fluorescence. 65°C is above the melting temperature of the nicked products (which will not be quenched, as shown at the right), but below the melting temperature of unnicked DNA (which will be quenched, as shown at the far left) [8]. The 5′ overhang in the initial substrate permits a nontagged version to be 32P-radiolabeled for gel-based assays.