Abstract
Piwi-interacting RNAs (piRNAs) were reported in 2006 as a novel class of small non-coding RNAs associated with Piwi proteins of the Argonaute/Piwi family. Recent studies have revealed not only the biogenesis of piRNAs and their roles in transposon silencing, but also the function of the Piwi-piRNA pathway in epigenetic and post-transcriptional regulation of gene expression. In addition, the function of this pathway in somatic cells has also been more systematically characterized. The new findings reveal the Piwi-piRNA pathway as a more general mechanism of gene regulation.
Introduction
Three major types of small RNAs, siRNAs, miRNAs, and piRNAs, associate with proteins in the Argonaute/Piwi family to execute sequence-specific gene silencing. Among the three types of small RNAs, piRNAs were discovered most recently and remain the least characterized. They are named by their exclusive association with Piwi but not Argonaute (Ago) subfamily proteins. Unlike the broad expression of siRNAs and miRNAs in most cell and tissue types, piRNAs are highly enriched in the germline. This expression pattern and the findings that mutations of piRNA biogenesis factors result in either germline stem cell loss or sterility of the mutant animals demonstrate the critical impact the Piwi-piRNA pathway exerts on germline development and functions [1–6]. Strong hypormophic mutations of Drosophila piwi result in reduced germline stem cell division and eventual germline stem cell loss [3,7]. Reduced level of Piwi homologs in Caenorhabditis elegans result in reduced germ cell division and reduced fertility [7,8]. Knockout mutations of mouse Piwi proteins Mili, Miwi, and Miwi2 result in male sterility in all cases, spermatogonial stem cell arrest and eventual loss in Mili and Miwi2 mutants, and meiotic arrest in Miwi mutant [4–6,9,10]. Genetic analysis of Piwi functions in the germline of these model organisms reinforced the critical role of Piwi proteins in germline development.
Numerous excellent reviews have detailed piRNA biogenesis pathway and contrasted it with miRNA and siRNA biogenesis [11–13]. Here we highlight some less discussed points. First, in contrast to the ~500 loci encoding miRNAs in most eukaryotic genomes, the number of piRNAs ranges up to 100 000's. Second, both the RNA polymerase transcribing the primary transcripts and the nuclease generating primary piRNAs from the primary transcripts are yet to be identified; an endonuclease Zucchini has been implicated in generating the 5′ end of piRNA [14,15]. Third, the mechanism of RNA target selection by the piRNA-induced silencing complex is unclear. Fourth, the mechanism by which PIWI-piRNA complexes regulate epigenetic processes via sequence specificity remains unclear.
The questions highlighted above remain unsolved partially because studies of piRNA functions have mostly been focused on transposon silencing in the germline. Indeed, mutations of genes involved in piRNA biogenesis result in increased transposition and DNA damage in germ cells [16,17] and because a subpopulation of piRNAs appears to suppress retrotransposon activities in the germline [18,19]. These studies, however, often overlook substantial evidence demonstrating that the Piwi-piRNA pathway significantly influences epigenetic mechanisms. Drosophila Piwi and its homolog Aubergine have been shown to affect transcriptional silencing [20], to be required for heterochromatin formation [21], and to affect Polycomb Group-mediated transgene silencing [22]. Furthermore, molecular and biochemical analyses of Piwi and piRNAs demonstrated that Piwi protein directly associates with and recruits HP1a [23,24], that the Piwi-piRNA complex directly binds to piRNA-complementary sequence in the genome and is required for proper histone modification of the sequence [25], and that maternally deposited piRNAs can epigenetically mediate transposon suppression in the offspring [26]. In addition, analyses of mouse Piwi homologs, Mili and Miwi2, indicated that they mediate DNA methylation in the male germline during embryogenesis [27–29]. Together, the above observations implicate epigenetic function of the Piwi-piRNA pathway and its importance for germline health and functions.
The Piwi-piRNA pathway in epigenetic regulation
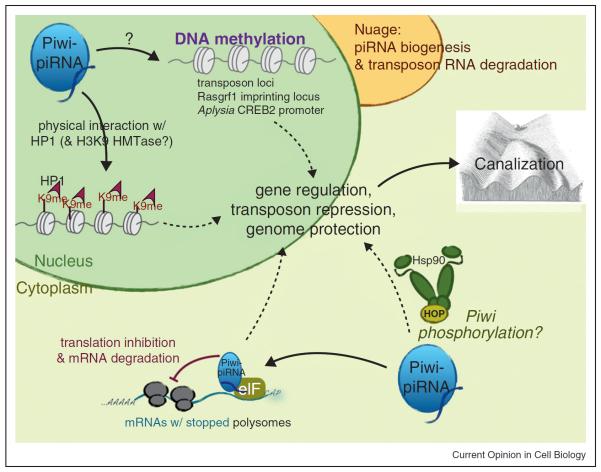
The Piwi-piRNA pathway can regulate epigenetic processes via the following mechanisms (Figure 1):
Figure 1.
The Piwi-piRNA pathway exhibits multiple regulatory functions of gene expression in diverse organisms. In the nucleus, PIWI-piRNA complex can regulate HP1, H3K9 methylation, and DNA methylation to influence transcription of selected protein-encoding genes, imprinting loci, or transposons. In the cytoplasm, PIWI-piRNA complex can suppress transposons in the nuage, interact with Hsp90-HOP to influence canalization, or interact with translational initiators (eIF) to inhibit polysomes and subsequent protein translation. This model is modified from Juliano et al. (2011) Ann. Rev. Genetics 45:447–69.
Histone modifications
Histone modifications are the predominant means by which epigenetic information is transmitted from parents to offspring. Drosophila Piwi protein has been shown to directly influence histone modifications at a piRNA target site [25] and to functionally associate with HP1a and histone H3 lysine9 methylation (H3K9me) [23,24]. HP1a and H3K9me are both characteristic marks of transcriptionally silenced chromatin. Recent works expanded on these observations by demonstrating that Piwi-piRNAs direct the genome-wide methylation of H3K9 to transcriptionally silence transposable elements in Drosophila ovarian somatic cells [30•] and that the molecular link between Piwi, HP1 and H3K9me may be evolutionarily conserved from Drosophila to C. elegans. The Piwi ortholog in C. elegans, PRG-1, can initiate trans-generational gene silencing in the germline by influencing H3K9me, HP1, and histone methyltransferases [31,32]. However, it is not clear whether PRG-1 initiates transcriptional silencing by physically associating with HP1 or by an alternative mechanism.
DNA methylation
DNA methylation is another epigenetic silencing mark functionally linked to Piwi. Mouse Piwi proteins Mili and Miwi2 are shown to regulate DNA methylation at transposon loci [27–29]. It seems that piRNAs can also direct DNA methylation on non-transposon loci, such as the Rasgrf1 locus in the mouse male germline to regulate genomic imprinting [33••] and the CREB2 promoter in Aplysia neurons to influence long-term memory plasticity [34•]. Even though the molecular mechanism by which piRNAs influence DNA methyltransferases is not clear, the high evolutionary conservation of this function is nevertheless remarkable.
The Piwi-piRNA pathway in canalization
Canalization was first described by Conrad Waddington in 1942 as the phenomenon in which `epigenetic landscape' canalizes genotypic variations into manifesting phenotypic similarities [35]. In modern terms, canalization is recognized as a molecular mechanism that buffers the impact of genotypic and environmental variations on phenotype to ensure developmental robustness. A few molecules have been identified for their role in canalization, among which the heat-shock protein 90 kDa (Hsp90) is the best characterized [36,37]. In addition, characterization of Hsp90 interactors showed that epigenetic factors interact with Hsp90 to facilitate canalization [38,39].
Small RNAs participate in canalization, as proved by the functional studies of miR-1 and miR-iab-4-5p in Drosophila. First is the demonstration that miR-1, a muscle-specific miRNA, impacts muscle differentiation only during the stressful rapid growth phase [40]. Second is the finding that miR-iab-4-5p mis-expression results in a haltere-to-wing transformation, a classic phenotype observed by Waddington [41] and shown to be canalized by Ubx, a molecular target of miR-iab-4-5p [42].
A recent work has linked canalization to the Piwi-piRNA pathway by demonstrating that Hsp90 influences piRNA production and transposon suppression, and that the mutation of Spn-E, a piRNA biogenesis factor, mediates canalization [43•]. Further investigation has shown that Piwi protein itself mediates canalization, that Hsp90, its accessory protein Hop, and Piwi form a complex to regulate canalization, and that Hsp90 is required for Piwi phosphorylation. These findings establish a molecular pathway in which Hsp90 and Hop mediate canalization via the Piwi-piRNA pathway, likely by regulating Piwi activity through phosphorylation [44••]. In addition, these studies imply that piRNAs in the germline or gametes contribute to canalization in the offspring. Hsp90 can influence multiple functions of the Piwi-piRNA pathway, which include canalization as well as transposon suppression (Figure 1). Significant work is needed to determine whether transposon mobilization directly influences canalization and to further reveal the mechanism by which the Hsp90-Hop-Piwi-piRNA pathway regulates canalization.
The Piwi-piRNA pathway in post-transcriptional and somatic regulation
Previous studies of the epigenetic regulatory functions of Drosophila Piwi indicated that it could influence epigenetic mechanisms outside the germline tissues [21,22,25,45]. However, the findings that animals containing piwi mutations or mutations in other genes for piRNA biogenesis manifest sterility but no other observed deleterious effects led to the presumption that Piwi-piRNAs do not significantly impact tissues outside the germline. These findings, however, cannot rule out the long-term impact of the Piwi-piRNA mechanisms in genomic stability and organismal health; nor can it rule out the role of Piwi-piRNA functions beyond the basic survival requirement under laboratory conditions.
A clear demonstration of the post-transcriptional (and somatic) function of the Piwi-piRNA mechanism is piRNA-mediated regulation of Nanos mRNA deadenylation and degradation in establishing the posterior Nanos gradient in the embryo [46••], which is critical for the establishment of anterior–posterior axis during early embryogenesis. Intriguingly, piRNAs and Piwi subfamily members Aubergine and Ago3 in Drosophila embryos physically associate with the CCR4-Smg complex, which dictates gradient localization of proteins that include Nanos and Hsp83/Hsp90 [47]. Therefore it is likely that the Piwi-piRNA complex can collaborate with the CCR4-Smg complex to mediate translational regulation of gene targets other than Nanos.
A clear line of evidence for the somatic function is the effect of piwi mutation in the Drosophila eye, a somatic organ far away from the gonad [23]. Another demonstration of the somatic function of the Piwi-piRNA mechanism is that piRNAs can influence the gene expression of CREB2 in Aplysia neurons to mediate long-term memory formation [34•]. This is a convincing case that the Piwi-piRNAs pathway can exhibit critical functions outside the germline tissues, even though it does not immediately impact the viability of the organism.
The piRNAs responsible for regulating DNA methylation of the CREB2 promoter in Aplysia neurons were identified by deep sequencing analysis as those that exhibit sequence complementarity to the CREB2 promoter [34•]. Two other reports also utilized deep sequencing analysis to identify piRNA-like species in hippocampal neurons [48•] and mammalian tissues that include cortex, epididymis, prostate, and the hematopoietic system [49•]. These identified small RNAs are piRNA-like because most of them exhibit the 5′ uridine signature of primary piRNAs [48•,49•], many exhibit the 10th adenosine signature of secondary piRNAs [49•], and significant populations (e.g. ~75 000 in hippocampal neurons) are identical to piRNAs bound by the mouse Piwi homolog Miwi in testis [48•]. One intriguing finding shared by all three studies is that these piRNA-like small RNAs are 10-fold to 100-fold less than the amount of piRNAs found in the germline, reflecting the possibility that piRNAs are of very low abundance or present in a very small number of cells in somatic tissues where they are expressed. Systematic characterization of piRNAs and Piwi proteins in tissues outside the germline could address these fascinating questions.
Concluding remark and future directions
Studies from the past few years have revealed the functions of the Piwi-piRNA pathway in regulating epigenetic and post-transcriptional gene regulation, in influencing somatic cell functions, and in mediating canalization to enhance developmental robustness. Because the Piwi-piRNA complex exerts its functions via piRNA sequence specificity, the high complexity of piRNAs allows them to act on a wide range of targets in the genome, and to generate broad impact on genome-wide gene regulation at multiple levels. Of the many fascinating questions that remain, a few stand out. How does the Piwi-piRNA complex function in the somatic cells outside the germline, with apparently few piRNA molecules present? How do Piwi-piRNA complexes interact with canonical epigenetic machinery in the nucleus to achieve epigenetic programming? And, whether/how do proteins interact with tens of thousands of different piRNAs in the cytoplasm to regulate gene expression? Answers to these questions will represent significant advancement in our understanding of the piRNA biology.
Acknowledgements
Work in the Lin lab is supported by NIH (DP1CA174418 and R37HD042012), the Connecticut Stem Cell Research Foundation, the G Harold and Leila Mathers Foundation, and the Ellison Medical Foundation. JCP is supported by an NIH Pathway to Independence (PI) Award (K99-HD071011).
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Cox DN, Chao A, Lin H. Piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez-Reyes A, Elliott H, St Johnston D. Oocyte determination and the origin of polarity in Drosophila: the role of the spindle genes. Development. 1997;124:4927–4937. doi: 10.1242/dev.124.24.4927. [DOI] [PubMed] [Google Scholar]
- 3.Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- 4.Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. Miwi2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Deng W, Lin H. Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 6.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 7.Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang G, Reinke V. A C. elegans piwi, prg-1, regulates 21u-RNAs during spermatogenesis. Curr Biol. 2008;18:861–867. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomson T, Lin H. The biogenesis and function of piwi proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H. Mili a piwi-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J Biol Chem. 2009;284:6507–6519. doi: 10.1074/jbc.M809104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saxe JP, Lin H. Small noncoding RNAs in the germline. Cold Spring Harbor Perspect Biol. 2011;3:a002717. doi: 10.1101/cshperspect.a002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gangaraju VK, Lin H. MicroRNAs key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czech B, Hannon GJ. Small RNA sorting: matchmaking for argonautes. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. The structural biochemistry of zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012;491:279–283. doi: 10.1038/nature11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, et al. Structure and function of zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491:284–287. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- 16.Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an atr/chk2 DNA damage response. Dev Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Pane A, Schupbach T. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol. 2007;17:637–642. doi: 10.1016/j.cub.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aravin AA, Hannon GJ, Brennecke J. The piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 19.O'Donnell KA, Boeke JD. Mighty piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and post-transcriptional transgene silencing in Drosophila. Mol Cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- 21.Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 22.Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi components are required for nuclear clustering of polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 23.Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila piwi associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendez DL, Kim D, Chruszcz M, Stephens GE, Minor W, Khorasanizadeh S, Elgin SC. The HP1a disordered c terminus and chromo shadow domain cooperate to select target peptide partners. Chembiochem. 2011;12:1084–1096. doi: 10.1002/cbic.201000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin H, Lin H. An epigenetic activation role of piwi and a piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 26.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate Mili in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 29.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, et al. DNA methylation of retrotransposon genes is regulated by piwi family members Mili and Miwi2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Sienski G, D̂nertas D, Brennecke J. Transcriptional silencing of transposons by piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]; Genome-wide profiles of the effect of Piwi and Maelstrom, a piRNA pathway effector, on histone H3 lysine 9 methylation and RNA Polymerase II on transposon loci.
- 31.Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr, Mello CC. PiRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, Pintacuda G, et al. PiRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, Gotoh K, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse rasgrf1 locus. Science. 2011;332:848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]; Influence by Piwi-piRNA on DNA methylation can exert beyond transposon loci and over an imprinting locus, which is critical for early embryogenesis.
- 34•.Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]; Influence by Piwi-piRNA on DNA methylation takes place in the nervous system to influence critical neural functions.
- 35.Waddington C. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. [Google Scholar]
- 36.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 37.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 38.Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 39.Tariq M, Nussbaumer U, Chen Y, Beisel C, Paro R. Trithorax requires hsp90 for maintenance of active chromatin at sites of gene expression. Proc Natl Acad Sci U S A. 2009;106:1157–1162. doi: 10.1073/pnas.0809669106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waddington CH. Genetic assimilation of the bithorax phenotype. Evol Int J Org Evol. 1956;10:1–13. [Google Scholar]
- 42.Ronshaugen M, Biemar F, Piel J, Levine M, Lai EC. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev. 2005;19:2947–2952. doi: 10.1101/gad.1372505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Specchia V, Piacentini L, Tritto P, Fanti L, D`Alessandro R, Palumbo G, Pimpinelli S, Bozzetti MP. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature. 2010;463:662–665. doi: 10.1038/nature08739. [DOI] [PubMed] [Google Scholar]; First demonstration that transposon silencing is connected to canalization.
- 44••.Gangaraju VK, Yin H, Weiner MM, Wang J, Huang XA, Lin H. Drosophila piwi functions in hsp90-mediated suppression of phenotypic variation. Nat Genet. 2011;43:153–158. doi: 10.1038/ng.743. [DOI] [PMC free article] [PubMed] [Google Scholar]; First molecular link between Hsp90, and the PIWI-piRNA pathway, the isolation of the first `canalization complex', and first systematic evaluation of the relative contribution of epigenetic suppression and transposon silencing mechanisms to canalization.
- 45.Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/jumonji coordinates control of prc2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early drosophila embryo. Nature. 2010;467:1128–1132. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]; Piwi-piRNA pathway regulates localization and expression of Nanos, which is required for early embryogenesis.
- 47.Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA. Smaug recruits the ccr4/pop2/not deadenylase complex to trigger maternal transcript localization in the early drosophila embryo. Curr Biol. 2005;15:284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- 48•.Lee EJ, Banerjee S, Zhou H, Jammalamadaka A, Arcila M, Manjunath BS, Kosik KS. Identification of piRNAs in the central nervous system. RNA. 2011;17:1090–1099. doi: 10.1261/rna.2565011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Isolation of piRNA-like species outside the germline.
- 49•.Yan Z, Hu HY, Jiang X, Maierhofer V, Neb E, He L, Hu Y, Hu H, Li N, Chen W, et al. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 2011;39:6596–6607. doi: 10.1093/nar/gkr298. [DOI] [PMC free article] [PubMed] [Google Scholar]; Isolation of piRNA-like species outside the germline.