Abstract
Cells acquire their fate in vivo in the context of a complex microenvironmental “niche” comprised of heterologous cell types, signaling molecules, extracellular matrix, biophysical forces, and metabolic substrates. Now Calderón and Boehm report the “refunctionalization” of a defective thymic epithelial niche, offering insights into how signaling molecules may be hierarchically organized to direct differentiation outcomes.
The acquisition of cell fate is a process that has fascinated biologists ever since “epigenesis” or “the unfolding of an organism” theory of development superseded the “preformation” theories in the 19th century. In the adult organism, the process of cell-fate determination plays out during the homeostasis of tissues with high rates of cell turnover. In these tissues, cell replenishment depends primarily on the division of multipotent stem or progenitor cells, instead of the division of mature cells. A major advantage of this model is that lineage choices are often ongoing, which allows the tissue to respond dynamically to physiologic challenge because the cell types generated can be modulated. The tissue that best exemplifies this dynamic multi-lineage determination in the adult is the blood.
Studying fate determination of the blood lineage has led to both biologic and medical advances of indisputable importance. The ability to produce red cells, granulocytes, and platelets by treatments with recombinant cytokines has changed the lives of countless individuals with cancer, kidney failure, and a host of other disorders. But it has been far more difficult to unravel what dictates lineage specification further upstream, such as at the level of multipotent progenitors or stem cells. The difficulty is primarily because cells in isolation, plus various protein additives, do not recapitulate the complex system of signals encountered in vivo. Indeed, Schofield proposed in 1978 that the controlled differentiation and self-renewal of stem cells take place in an anatomically definable site, called the microenvironmental “niche” of stem cells (Schofield, 1978). Understanding the molecular and cellular components of these niches has become an area of intense investigation. Now Calderón and Boehm (2012) present an elegant study that creatively deconvolutes the signaling required to build a tissue niche, not for stem cells but rather for progenitors, and not in the bone marrow but rather in the thymus. To uncover this niche recipe, Calderón and Boehm cleverly exploit a defect of epithelial cells, which are known to play a role in thymic control of T cell generation.
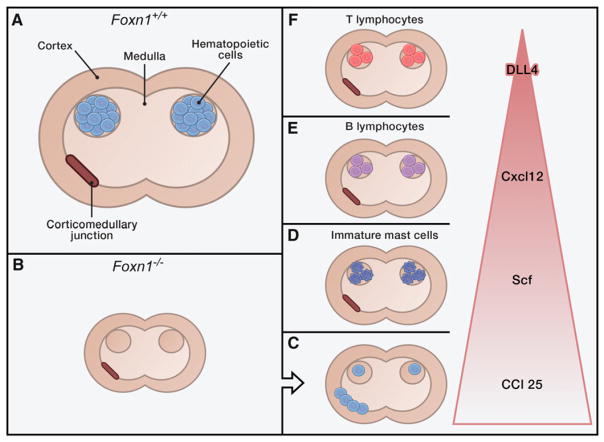
In all jawed vertebrates, thymic epithelial cells (TECs) depend upon the expression of the forkhead transcription factor Foxn1 (Nehls et al., 1994, 1996). Mice deficient in Foxn1 are nude mice that are defective in keratin production; these mice are hairless, but they are also immunodeficient due to a complete loss of intrathymic T cell development. A normal thymus consists of two major parts: the cortex, where T cell receptor rearrangements and positive selection of immature thymocytes occur, and a medulla, which is responsible for negative selection of T cells with self-antigens. Both regions depend upon TECs to perform their tasks, but the TECs in the medulla are distinct in that they their function depends on the expression of the transcription factor AIRE (Anderson et al., 2002). The cortico-medullary junction is the site at which hematopoietic progenitor cells enter the adult thymus. After this, these progenitor cells are known to encounter a number of extracellular signals that are important for regulating their differentiation (reviewed in Rodewald, 2008). However, it has been extremely difficult to discern exactly how these factors combine together to support hematopoietic progenitor cells and orchestrate cell-fate outcomes.
Exploiting the dysfunctional TECs of nude mice, Calderón and Boehm engineer mice that selectively express extracellular signaling molecules known to be important in T cell development. These molecules are not expressed in the nude TECs, so the authors add them back by creating transgenic mice in which the Foxn1 promoter drives the signaling compound of interest. In this way, Calderón and Boehm test the ability of specific molecules or their combinations to refunctionalize the thymus. The molecules they evaluate are the chemokines Ccl25 and Cxcl12, the cytokine Scf (stem cell factor), and the Notch ligand DLL4.
Calderón and Boehm find that the number of hematopoietic cells (indicated by the pan-hematopoietic cell antigen CD45) modestly increases in the presence of either Ccl25, Cxcl12, or Scf but not the Notch ligand DLL4. This suggests that Cxcl12, Ccl25, and Scf are involved in localizing the hematopoetic cells. Depending on which factor is re-expressed, the cellular composition in the rudimentary thymus varies; specifically, the presence of Ccl25 increases lymphoid cells, Cxcl12 increases primarily B lineage-committed lymphoid cells, and Scf increases a population without clear lineage specification that appear to be myeloid progenitors. Intriguingly, combining at least two factors altered the composition and pattern of the accumulating thymic hematopoietic cells more profoundly than single factors alone. Simultaneous expression of Ccl25 and Scf creates a microenvironment particularly favorable for immature mast cells, a rather remarkable conversion to myelopoietic support for an organ regarded as a prototypical lymphoid niche. Adding Cxcl12 to Ccl25 and Scf changes the supportive landscape sufficiently that now CD19+ B lineage cells preferentially grow. These results argue that Cxcl12 is hierachically superior to Ccl25 and Scf for nurturing hemopoeitic lineages and is capable of converting a mast cell-favoring environment to a microenvironment promoting B cell development.
In subsequent experiments, Calderón and Boehm show that the Notch ligand DLL4 is situated at the top of the functional hierarchy of the four studied factors (Figure 1), with Cxcl12 promoting B cell development in the absence of the Notch ligand DLL4 but fostering T cell development in the presence of DLL4. The authors then demonstrate that expression of Cxcl12 and DLL4 alone are sufficient to support T cell development to the CD4/CD8 double-positive stage and that Ccl25 is at the foot of the pyramid of factors regulating T cell development in the thymus.
Figure 1. Recipe for Refunctionalizing a Thymic Niche.
(A) Simplified schema of a normal thymus in a mouse expressing the transcription factor Foxn1.
(B) The same schema for a thymus of a nude mouse that does not express Foxn1; here the thymus lacks hematopoietic elements.
(C–F) By re-expressing individual signaling factors in the thymus of the nude mice—Ccl25 (C), Scf (D), Cxcl12 (E), and DLL4 (F)—Calderón and Boehm (2012) uncover a hierarchical restructuring of the thymus. The combined expression of factors leads to phenotypes as described in this Preview.
Calderón and Boehm have thus developed a method of teasing apart the interactive relationships that comprise a cell-fate-determining microenvironment. They have done so in a way that highlights the importance of moving beyond reductionist approaches. Manipulating individual factors at one time may be helpful in identifying the participating molecules and even cell types, but this strategy gives little insight into, and perhaps may provide misleading information about, how a complex regulatory tissue functions physiologically. It is no surprise that combinatorial signals matter in defining cell state. This new study by Calderón and Boehm teaches that such signals can begin to be weighted and scored in a hierarchical order based on performance in functioning tissue in mammals in vivo. Combining such approaches with in-depth analysis of the intracellular events that accompany different cell-fate outcomes will ultimately give us the map for how cell identity is forged. That may enable us to become true cell engineers, perhaps even engineers of epigenesis.
References
- Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Mathis D. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- Calderón L, Boehm T. Cell. 2012;149:159–172. doi: 10.1016/j.cell.2012.01.049. this issue. [DOI] [PubMed] [Google Scholar]
- Nehls M, Pfeifer D, Schorpp M, Hedrich H, Boehm T. Nature. 1994;372:103–107. doi: 10.1038/372103a0. [DOI] [PubMed] [Google Scholar]
- Nehls M, Kyewski B, Messerle M, Waldschütz R, Schüddekopf K, Smith AJ, Boehm T. Science. 1996;272:886–889. doi: 10.1126/science.272.5263.886. [DOI] [PubMed] [Google Scholar]
- Rodewald HR. Annu Rev Immunol. 2008;26:355–388. doi: 10.1146/annurev.immunol.26.021607.090408. [DOI] [PubMed] [Google Scholar]
- Schofield R. Blood Cells. 1978;4:7–25. [PubMed] [Google Scholar]