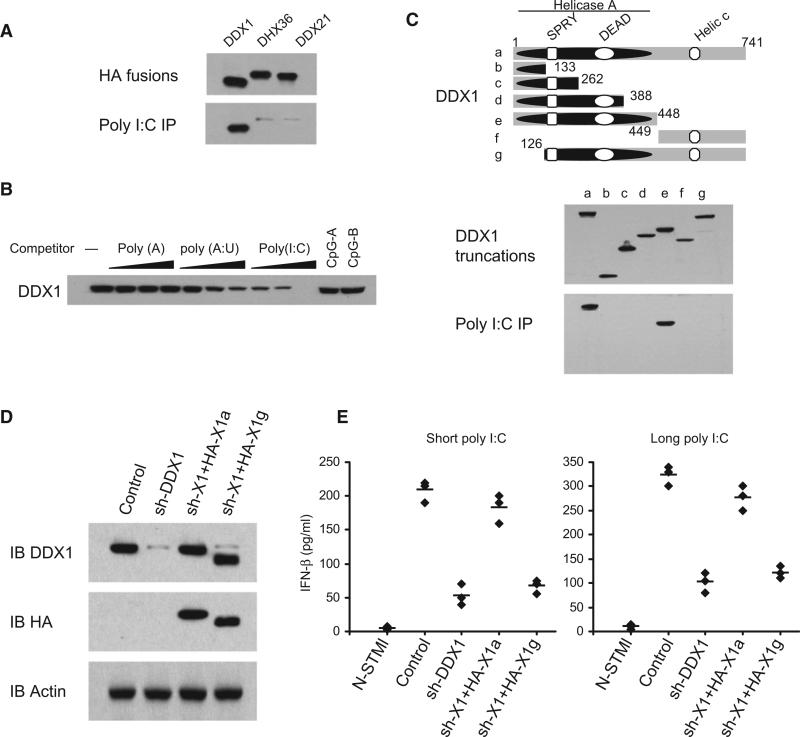
Figure 2. The Helicase A Domain of DDX1 Binds Poly I:C.
(A) Pull-down assays were performed by incubating purified HA-DDX1, HA-DHX36, or HA-DDX21 with poly I:C-biotin-NA beads. Bound proteins were analyzed by immunoblotting with anti-HA.
(B) The mixture of HA-DDX1 and poly I:C-biotin-NA beads were incubated without polynucleotides or with polynucleotides at 0.5, 5, or 50 μg/ml concentration. Bound proteins were analyzed by immunoblotting with anti-HA.
(C) Schematic representations of DDX1 and its serial deletion mutants: (a) DDX1 full size, (b) DDX1 dSPRY, (c) DDX1 dDEAD, (d) DDX1 nDEAD, (e) DDX1 nHELICa, (f) DDX1 cHELICc, and (g) DDX1 SPY. Helicase A, Helicase ATP-binding domain; DEAD, Asp-Glu-Ala-Asp box motif; SPRY, SPla and the RYanodine domain; and Helic C, helicase C-terminal domain. Numbers denote amino acid residues. Truncated HA fusions were purified (middle panel) and individually incubated with poly I:C-biotin along with NA beads. Bound proteins were analyzed by immunoblotting with anti-HA (lower panel).
(D) Immunoblot of endogenous DDX1, recombinant HA-DDX1a and HA-DDX1g in D2SC with the indicated antibodies. D2SC treated with scrambled shRNA (control) or with shRNA targeting DDX1 (sh-DDX1). The sh-DDX1 cells were then transfected with HA-DDX1a (sh-X1+HA-X1a) or HA-DDX1g (sh-X1+HA-X1g) expression plasmids.
(E) ELISA of IFN-β production by indicated cells stimulated with short or long poly I:C delivered with Lipofectamine 2000 for 16 hr. Individual diamond represents the value from each independent experiment. Error bars represent the average value from at least three independent experiments.